|
|
1 an în urmă | |
|---|---|---|
| R | 1 an în urmă | |
| docker | 3 ani în urmă | |
| img | 2 ani în urmă | |
| inst | 3 ani în urmă | |
| man | 3 ani în urmă | |
| scripts | 2 ani în urmă | |
| src | 3 ani în urmă | |
| tests | 2 ani în urmă | |
| .gitattributes | 3 ani în urmă | |
| .gitignore | 2 ani în urmă | |
| DESCRIPTION | 2 ani în urmă | |
| LICENSE | 3 ani în urmă | |
| NAMESPACE | 3 ani în urmă | |
| README.md | 2 ani în urmă |
README.md
CALDER user manual
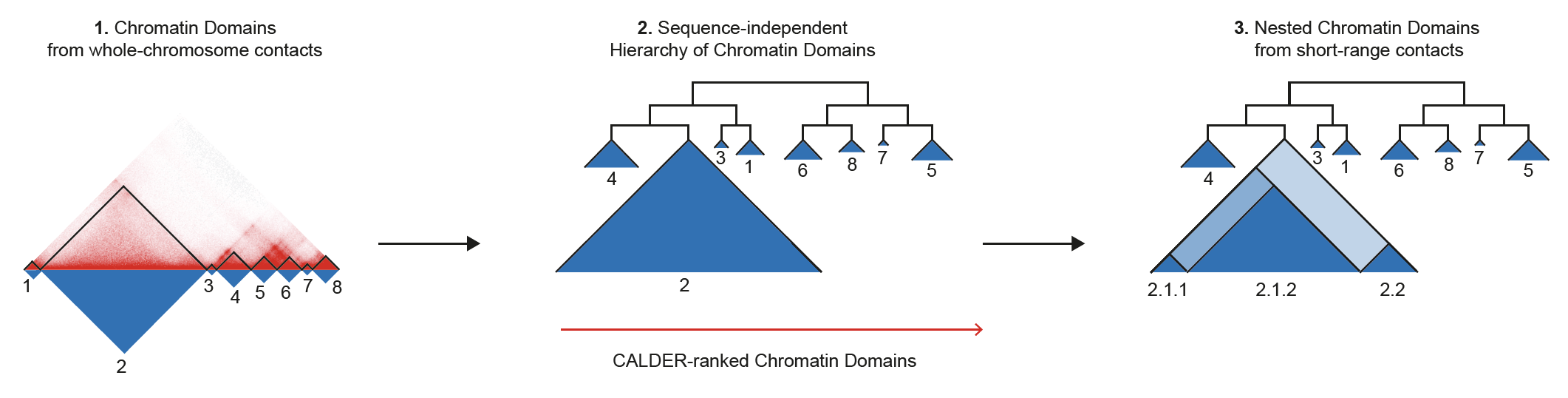
CALDER is a Hi-C analysis tool that allows: (1) compute chromatin domains from whole chromosome contacts; (2) derive their non-linear hierarchical organization and obtain sub-compartments; (3) compute nested sub-domains within each chromatin domain from short-range contacts. CALDER is currently implemented in R.
(A note on the performance of Calder vs PC-based approach)
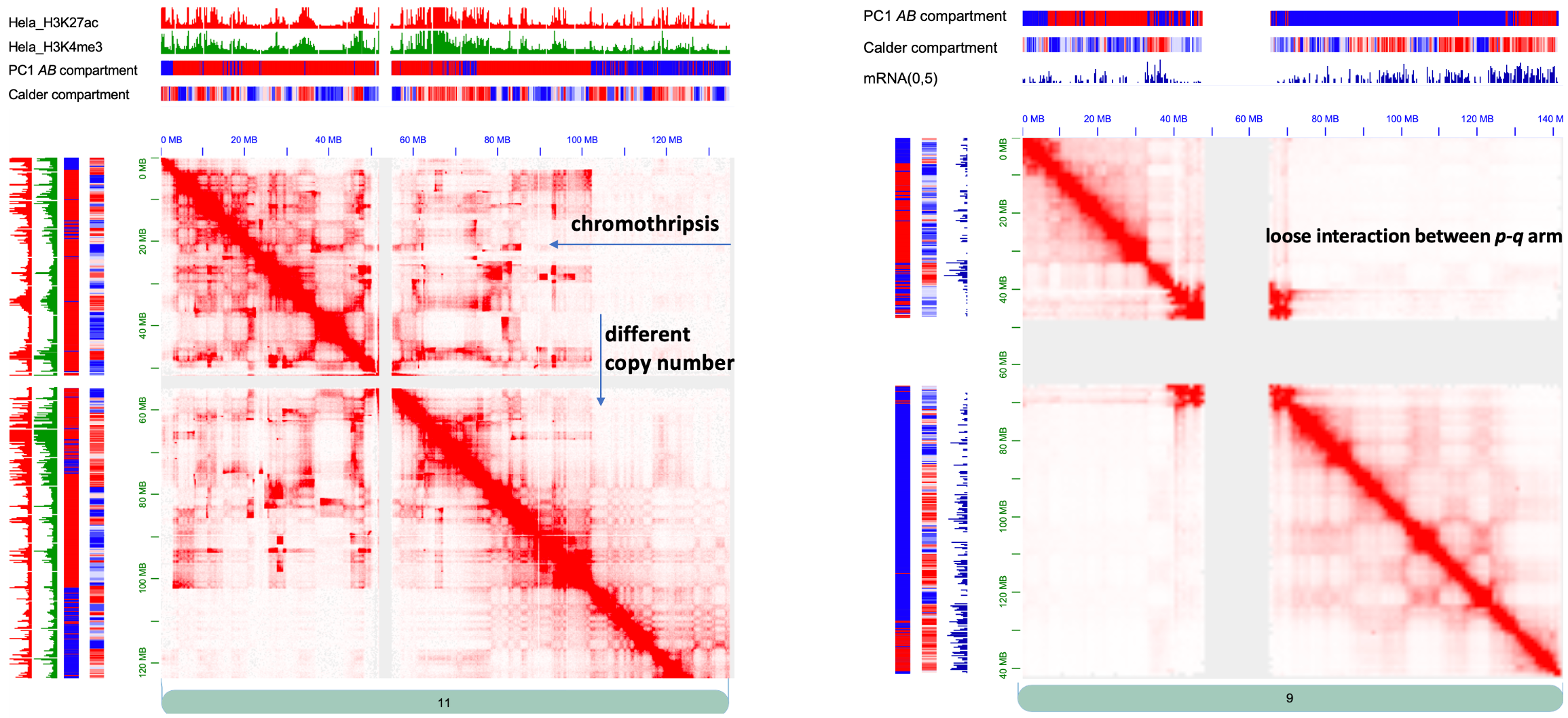
- PC1 (sometimes PC2) of the correlation matrix was typically used to define A/B compartment. We found Calder demonstrates superior robustness over PC-based approach in identifying meaningful compartments, particularly when faced with complex chromosomal structural variations (figure on the left) or loose interaction between the p and q arms (figure on the right)
Multiple new features were added in version 2.0
- Support for hg19, hg38, mm9, mm10 and other genomes
- Support input in .hic format generated by Juicer tools (https://github.com/aidenlab/juicer)
- Optimized bin_size selection for more reliable compartment identification
- Aggregated all chromosome output into a single file for easier visualization in IGV
- Added output in tabular .txt format at bin level for easier downstream analysis
Below we introduce two main updates:
(1) Optimized bin_size selection
Due to reasons such as low data quality or large scale structural variation, compartments can be unreliably called at one bin_size (equivalent to resolution in the literature) but properly called at another bin_size. We added an optimized bin_size selection strategy to call reliable compartments. This strategy is based on the observation from our large scale compartment analysis (https://www.nature.com/articles/s41467-021-22666-3), that although compartments can change between different conditions, their overall correlation cor(compartment_rank_1, compartment_rank_2) is high (> 0.4).
The strategy: given a bin_size specified by user, we call compartments with extended bin_sizes and choose the smallest bin_size such that no bigger bin_size can increase the compartment correlation with a reference compartment more than 0.05. For example, if correlation for bin_size=10000 is 0.2 while for bin_size=50000 is 0.6, we are more confident that the latter is more reliable; if correlation for bin_size=10000 is 0.5 while for bin_size=50000 is 0.52, we would choose the former as it has higher resolution.
bin_size is extended in the following way thus contact matrices at any larger bin_sizes can be aggregated from the input contact matrices directly:
if(bin_size==5E3) bin_sizes = c(5E3, 10E3, 50E3, 100E3)
if(bin_size==10E3) bin_sizes = c(10E3, 50E3, 100E3)
if(bin_size==20E3) bin_sizes = c(20E3, 40E3, 100E3)
if(bin_size==25E3) bin_sizes = c(25E3, 50E3, 100E3)
if(bin_size==40E3) bin_sizes = c(40E3, 80E3)
if(bin_size==50E3) bin_sizes = c(50E3, 100E3)
Note that this strategy is currently only available for hg19, hg38, mm9 and mm10 genome for which we generated high quality reference compartments using Hi-C data from: GSE63525 for hg19, https://data.4dnucleome.org/files-processed/4DNFI1UEG1HD for hg38, GSM3959427 for mm9, http://hicfiles.s3.amazonaws.com/external/bonev/CN_mapq30.hic for mm10.
(2) Support for other genomes
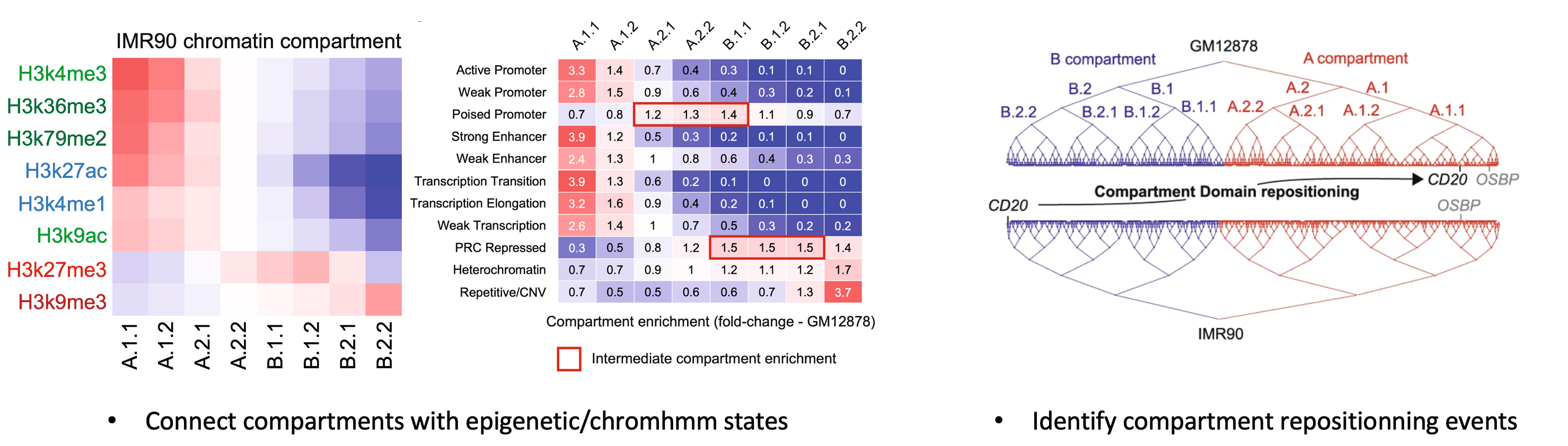
Although CALDER was mainly tested on human and mouse dataset, it can be applied to dataset from other genomes. One additional information is required in such case: a feature_track presumably positively correlated with compartment score (thus higher values in A than in B compartment). This information will be used for correctly determining the A/B direction. Some suggested tracks are gene density, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3 (or negative transform of H3K9me3) signals. Note that this information will not alter the hierarchical compartment/TAD structure, and can come from any external study with matched genome. An example of feature_track is given in the Usage section.
Installation
Installing from conda
The easiest way to get the package is to install from Bioconda:
conda install --channel bioconda r-calder2
Installing from source
Make sure all dependencies have been installed:
- R.utils (>= 2.9.0),
- doParallel (>= 1.0.15),
- ape (>= 5.3),
- dendextend (>= 1.12.0),
- fitdistrplus (>= 1.0.14),
- igraph (>= 1.2.4.1),
- Matrix (>= 1.2.17),
- rARPACK (>= 0.11.0),
- factoextra (>= 1.0.5),
- data.table (>= 1.12.2),
- fields (>= 9.8.3),
- GenomicRanges (>= 1.36.0)
- ggplot2 (>= 3.3.5)
- strawr (>= 0.0.9)
Clone its repository and install it from source:
On the command line:
git clone https://github.com/CSOgroup/CALDER2.git
cd CALDER2
Then, once inside of the R interpreter:
install.packages(".", repos = NULL, type="source") # install from the cloned source file
Install CALDER and dependencies automaticly:
One can also install directly from Github, together with the dependencies as follows:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("GenomicRanges")
install.packages("remotes")
remotes::install_github("CSOgroup/CALDER2.0")
Please contact yliueagle@googlemail.com for any questions about installation.
Use as a docker container
We provide a Docker image complete with all dependencies to run CALDER workflows.
# Pull the docker image from Dockerhub
docker pull lucananni93/calder2
# Run the image
docker run -it lucananni93/calder2
# Once inside the image we can run the command line Calder tool
calder [options]
# or we can just enter R
R
# and load Calder
library(CALDER)
Usage
CALDER contains three modules: (1) compute chromatin domains; (2) derive their hierarchical organization and obtain sub-compartments; (3) compute nested sub-domains within each compartment domain.
Input data format
CALDER works on contact matrices compatible with that generated by Juicer tools (https://github.com/aidenlab/juicer), either a .hic file, or three-column dump table retrieved by the juicer dump (or straw) command (https://github.com/aidenlab/juicer/wiki/Data-Extraction):
16050000 16050000 10106.306
16050000 16060000 2259.247
16060000 16060000 7748.551
16050000 16070000 1251.3663
16060000 16070000 4456.1245
16070000 16070000 4211.7393
16050000 16080000 522.0705
16060000 16080000 983.1761
16070000 16080000 1996.749
...
feature_track should be a data.frame or data.table of 4 columns (chr, start, end, score), and can be generated directly from conventional format such as bed or wig, see the example:
library(rtracklayer)
feature_track = import('ENCFF934YOE.bigWig') ## from ENCODE https://www.encodeproject.org/files/ENCFF934YOE/@@download/ENCFF934YOE.bigWig
feature_track = data.table::as.data.table(feature_track)[, c(1:3, 6)]
> feature_track
chr start end score
chr1 534179 534353 2.80512
chr1 534354 572399 0
chr1 572400 572574 2.80512
chr1 572575 628400 0
... ... ... ...
chrY 59031457 59032403 0
chrY 59032404 59032413 0.92023
chrY 59032414 59032415 0.96625
chrY 59032416 59032456 0.92023
chrY 59032457 59032578 0.78875
Example usage (1): use contact matrix file in dump format as input
chrs = c(21:22)
## demo contact matrices in dump format
contact_file_dump = as.list(system.file("extdata", sprintf("mat_chr%s_10kb_ob.txt.gz", chrs),
package='CALDER'))
names(contact_file_dump) = chrs
## Run CALDER to compute compartments but not nested sub-domains
CALDER(contact_file_dump=contact_file_dump,
chrs=chrs,
bin_size=10E3,
genome='hg19',
save_dir=save_dir,
save_intermediate_data=FALSE,
n_cores=2,
sub_domains=FALSE)
## Run CALDER to compute compartments and nested sub-domains / will take more time
CALDER(contact_file_dump=contact_file_dump,
chrs=chrs,
bin_size=10E3,
genome='hg19',
save_dir=save_dir,
save_intermediate_data=TRUE,
n_cores=2,
sub_domains=TRUE)
Example (2): use contact matrices stored in an R list
chrs = c(21:22)
contact_file_dump = as.list(system.file("extdata", sprintf("mat_chr%s_10kb_ob.txt.gz", chrs),
package='CALDER'))
names(contact_file_dump) = chrs
contact_tab_dump = lapply(contact_file_dump, data.table::fread)
CALDER(contact_tab_dump=contact_tab_dump,
chrs=chrs,
bin_size=10E3,
genome='hg19',
save_dir=save_dir,
save_intermediate_data=FALSE,
n_cores=2,
sub_domains=FALSE)
Example (3): use .hic file as input
chrs = c(21:22)
hic_file = 'HMEC_combined_30.hic' ## can be downloaded from https://ftp.ncbi.nlm.nih.gov/geo/series/GSE63nnn/GSE63525/suppl/GSE63525_HMEC_combined_30.hic
CALDER(contact_file_hic=hic_file,
chrs=chrs,
bin_size=10E3,
genome='hg19',
save_dir=save_dir,
save_intermediate_data=FALSE,
n_cores=2,
sub_domains=FALSE)
Example (4): run CALDER on other genomes
## prepare feature_track
library(rtracklayer)
feature_track_raw = import('ENCFF934YOE.bigWig') ## from ENCODE https://www.encodeproject.org/files/ENCFF934YOE/@@download/ENCFF934YOE.bigWig
feature_track = data.table::as.data.table(feature_track_raw)[, c(1:3, 6)]
## Run CALDER
chrs = c(21:22)
contact_file_dump = as.list(system.file("extdata", sprintf("mat_chr%s_10kb_ob.txt.gz", chrs),
package='CALDER'))
names(contact_file_dump) = chrs
CALDER(contact_file_dump=contact_file_dump,
chrs=chrs,
bin_size=10E3,
genome='others',
save_dir=save_dir,
feature_track=feature_track,
save_intermediate_data=FALSE,
n_cores=2,
sub_domains=FALSE)
Parameters:
| Name | Description |
| --------------------- | ----------------------- |
| chrs | A vector of chromosome names to be analyzed, with or without 'chr'. Chromosome names should be consistent with those in contact_file_hic and feature_track if such files are provided
| contact_file_dump |A list of contact files in dump format, named by chrs. Each contact file stores the contact information of the corresponding chr. Only one of contact_file_dump, contact_tab_dump, contact_file_hic should be provided
| contact_tab_dump | A list of contact table in dump format, named by chrs, stored as an R object. Only one of contact_file_dump, contact_tab_dump, contact_file_hic should be provided
| contact_file_hic | A hic file generated by Juicer tools. It should contain all chromosomes in chrs. Only one of contact_file_dump, contact_tab_dump, contact_file_hic should be provided
| genome | One of 'hg19', 'hg38', 'mm9', 'mm10', 'others' (default). These compartments will be used as reference compartments for optimized bin_size selection. If genome = 'others', a feature_track should be provided (see below) and no optimized bin_size selection will be performed
| save_dir | the directory to be created for saving outputs
| bin_size | The bin_size (resolution) to run CALDER. bin_size should be consistent with the data resolution in contact_file_dump or contact_tab_dump if these files are provided as input, otherwise bin_size should exist in contact_file_hic. Recommended bin_size is between 10000 to 100000
| single_binsize_only | logical. If TRUE, CALDER will compute compartments only using the bin_size specified by the user and not do bin size optimization
| feature_track | A genomic feature track in data.frame or data.table format with 4 columns (chr, start, end, score). This track will be used for determining the A/B compartment direction when genome='others' and should presumably have higher values in A than in B compartment. Some suggested tracks can be gene density, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3 (or negative transform of H3K9me3 signals)
| save_intermediate_data | logical. If TRUE, an intermediate_data will be saved. This file can be used for computing nested sub-domains later on
| n_cores | integer. Number of cores to be registered for running CALDER in parallel
| sub_domains | logical, whether to compute nested sub-domains
Output Structure
The output of the workflow is stored in the folder specified by save_dir and will look like this:
save_dir/
|-- sub_domains
| `-- all_nested_boundaries.bed
|-- sub_compartments
| |-- cor_with_ref.txt
| |-- cor_with_ref.pdf
| |-- cor_with_ref.ALL.txt
| |-- all_sub_compartments.tsv
| `-- all_sub_compartments.bed
`-- intermediate_data
|-- sub_domains
| |-- chr22_sub_domains_log.txt
| |-- chr22_nested_boundaries.bed
| |-- chr21_sub_domains_log.txt
| `-- chr21_nested_boundaries.bed
`-- sub_compartments
|-- 50kb
| |-- chr22_sub_compartments.bed
| |-- chr22_log.txt
| |-- chr22_domain_hierachy.tsv
| |-- chr22_domain_boundaries.bed
| |-- chr21_sub_compartments.bed
| |-- chr21_log.txt
| |-- chr21_domain_hierachy.tsv
| `-- chr21_domain_boundaries.bed
|-- 10kb
| |-- chr22_sub_compartments.bed
| |-- chr22_log.txt
| |-- chr22_intermediate_data.Rds
| |-- chr22_domain_hierachy.tsv
| |-- chr22_domain_boundaries.bed
| |-- chr21_sub_compartments.bed
| |-- chr21_log.txt
| |-- chr21_intermediate_data.Rds
| |-- chr21_domain_hierachy.tsv
| `-- chr21_domain_boundaries.bed
`-- 100kb
|-- chr22_sub_compartments.bed
|-- chr22_log.txt
|-- chr22_domain_hierachy.tsv
|-- chr22_domain_boundaries.bed
|-- chr21_sub_compartments.bed
|-- chr21_log.txt
|-- chr21_domain_hierachy.tsv
`-- chr21_domain_boundaries.bed
File description:
- The
sub_domainsfolder stores the nested boundaries - The
sub_compartmentsfolder stores the optimal compartment information, including files:
| Name | Description |
| --------------------- | ----------------------- |
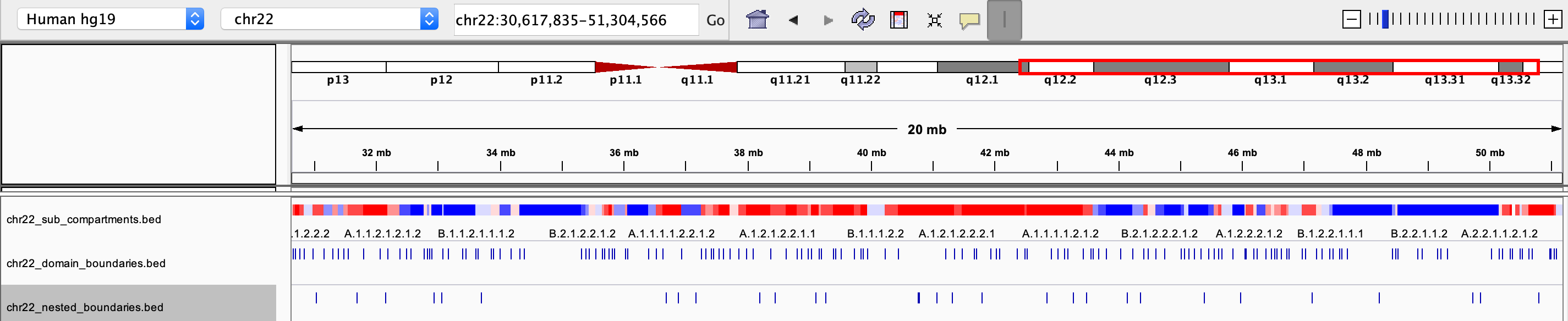
| all_sub_compartments.bed | a .bed file containing the optimal compartments for all chrs, that can be visualized in IGV. Different colors were used to distinguish compartments (at the resolution of 8 sub-compartments)
| all_sub_compartments.tsv | optimal compartments stored in tabular text format. Each row represents one 10kb region
| cor_with_ref.ALL.txt | a plot of correlation between compartment rank and the reference compartment rank for each of extended bin_sizes, and the optimal bin_size that is finally selected
| cor_with_ref.pdf | correlation of compartment rank with the reference compartment rank using the optimal bin_size
- The
intermediate_datafolder stores information at each of the extendedbin_sizesfor each chromosome
| Name | Description |
| --------------------- | ----------------------- |
| chrxx_domain_hierachy.tsv | information of compartment domain and their hierarchical organization. The hierarchical structure is fully represented by compartment_label, for example, B.2.2.2 and B.2.2.1 are two sub-branches of B.2.2. The pos_end column specifies all compartment domain borders, except when it is marked as gap, which indicates it is the border of a gap chromosome region that has too few contacts and was excluded from the analysis (e.g., due to low mappability, deletion, technique flaw)
| chrxx_sub_compartments.bed | a .bed file containing the compartment information, that can be visualized in IGV. Different colors were used to distinguish compartments (at the resolution of 8 sub-compartments)
| chrxx_domain_boundaries.bed | a .bed file containing the chromatin domains boundaries, that can be visualized in IGV
| chrxx_nested_boundaries.bed | a .bed file containing the nested sub-domain boundaries, that can be visualized in IGV
| chrxx_intermediate_data.Rds | an Rds file storing the intermediate_data that can be used to compute nested sub-domains
| chrxx_log.txt, chrxx_sub_domains_log.txt | log files storing the status and running time of each step
All .bed files can be view directly through IGV:
Running time:
For the computational requirement, running CALDER on the GM12878 Hi-C dataset at bin size of 40kb took 36 minutes to derive the chromatin domains and their hierarchy for all chromosomes (i.e., CALDER Step1 and Step2); 13 minutes to derive the nested sub-domains (i.e., CALDER Step3). At the bin size of 10kb, it took 1 h 44 minutes and 55 minutes correspondingly (server information: 40 cores, 64GB Ram, Intel(R) Xeon(R) Silver 4210 CPU @ 2.20GHz). The evaluation was done using a single core although CALDER can be run in a parallel manner.
Citation
If you use CALDER in your work, please cite: https://www.nature.com/articles/s41467-021-22666-3
Contact information
- Author: Yuanlong LIU
- Affiliation: Computational Systems Oncology group, Department of Computational Biology, University of Lausanne, Switzerland
- Email: yliueagle@googlemail.com