|
|
@@ -1,61 +1,62 @@
|
|
|
Creating Content Types
|
|
|
======================
|
|
|
|
|
|
- Tripal v3 comes with some pre-defined content types, however you have the ability to create new Content Types through the Administrative user interface! In Tripal v3, all content types are defined by public ontology terms. This has a number of advantages:
|
|
|
-
|
|
|
-1. Facilitates sharing between Tripal sites,
|
|
|
-2. Provides a clear indication of what content is available in your site,
|
|
|
-3. Makes content creation more intuitive (add a "Gene" rather then a "feature")
|
|
|
-4. Allows complete customization of what data types your site provides
|
|
|
-5. Integrates tightly with web services allowing Tripal to adhere to RDF specifications
|
|
|
-
|
|
|
-Find an Ontology Term
|
|
|
----------------------
|
|
|
-This is likely the most difficult step as it seems ontologies are both plentiful and sparse at the same time. We highly recommend discussing your content with the community so that the same term can be used across Tripal sites for the same data. If your data is genomics based, you need look no further than the sequence ontology. The following table lists some recommended terms for common genomic data:
|
|
|
-
|
|
|
-.. csv-table::
|
|
|
- :header: "Term Name", "Accession", "Examples", "Comments"
|
|
|
-
|
|
|
- "genome", "SO:0001026", "specific genome assembly", "*See the note below about genome assemblies.*"
|
|
|
- "chromosome", "SO:0000340", "", "Think carefully about whether it's useful to provide pages for chromosomes. The typical fields provided for content stored in the Chado feature table such as relationship and sequence listing can cause long page loads for not very useful information. Thus if you are going to provide pages you might want to include more summary fields based on materialized views to provide more useful content to users."
|
|
|
- "supercontig", "SO:0000148", "", "This term is a synonym for scaffold and should be used for such"
|
|
|
- "contig", "SO:0000149", "", ""
|
|
|
- "gene", "SO:0000704", "", ""
|
|
|
- "mRNA", "SO:0000234", "", ""
|
|
|
- "sequence_variant", "SO:0001060", "SNP, MNP, Indel", "We recommend grouping all variants into a single content type in order to provide a common page layout across variant types and to make searching across variants (e.g. show me all variants in a given region) easier. This also makes it easier to support new variant types in the future."
|
|
|
- "genetic_marker", "SO:0001645", "KASP, Exome Capture, GBS", "We recommend grouping all genetic markers for the same reasons we recommend grouping all sequence variants. You can then specify the type of variant or marker in the featureprop table."
|
|
|
+Tripal v3 comes with some pre-defined content types, however you have the ability to create new Content Types through the Administrative user interface! In Tripal v3, all content types are defined by `Controlled Vocabulary (CV) terms <https://en.wikipedia.org/wiki/Controlled_vocabulary>`_. This has a number of advantages:
|
|
|
|
|
|
-.. note::
|
|
|
+1. Facilitates sharing between Tripal sites.
|
|
|
+2. Provides a clear indication of what content is available on your site.
|
|
|
+3. Makes content creation more intuitive from Tripal v2 (add a "Gene" rather then a "feature").
|
|
|
+4. Allows complete customization of what data types your site provides.
|
|
|
+5. Integrates tightly with web services allowing Tripal to adhere to RDF specifications.
|
|
|
+
|
|
|
+Find a Controlled Vocabulary (CV) Term
|
|
|
+---------------------------------------
|
|
|
+
|
|
|
+Before creating a new content type for your site you must identify a CV term that best matches the content type you would like to create. CVs are plentiful and at times selection of the correct term from the right vocabulary can be challenging. If there is any doubt about what term to use, then it is best practice to reach out to others to confirm your selection. The Tripal User community is a great place to do this by posting a description of your content type and your proposed term on the `Tripal Issue Queue <https://github.com/tripal/tripal/issues>`_. Confirming your term with others will also encourage re-use across Tripal sites and improve data exchagnge capabilities.
|
|
|
|
|
|
- There are a few ways you might choose to store genome assemblies in Chado:
|
|
|
+The `EBI's Ontology Lookup Service <http://www.ebi.ac.uk/ols/index>`_ is a great place to locate terms from public vocabularies. At this site you can search for terms for your content type. If you can not find an appropriate term in a public vocabulary or via discussion with others then you create a new **local** term within the **local** vocabulary that comes with Tripal.
|
|
|
|
|
|
- 1. The analysis table. Chromosomes or scaffolds are linked via the analysisfeature table. This is the method Tripal uses by default.
|
|
|
- 2. The feature table. Chromosomes or scaffolds are also stored in the feature table and linked to the genome feature via the feature_relationship table.
|
|
|
- 3. The project table. Chromosomes or scaffolds are stored in the feature table and linked using the project_feature table.
|
|
|
- There is discussion in the community about which of these three is the best method.
|
|
|
+.. warning::
|
|
|
|
|
|
-For non-genomic data, it can be challenging to find an appropriate term for your content type as both the number and maturity of ontologies is decreased. However, a good starting place is `EBI's Ontology Lookup Service <http://www.ebi.ac.uk/ols/index>`_. At this site you can search for terms that may match your content type. You may find a perfect term, but if you are uncertain if a term is appropriate please open an issue on `Tripal's GitHub project page <https://github.com/tripal/tripal/issues>`_ to encourage discussion among the community for the ideal term. Not only will this provide you with help and input in your own search for an ontology term, it will also help others with similar data. We intend to add to this page once decisions are made for other data types to make this process easier and encourage sharing of data between Tripal sites. If all else fails and you can not find an appropriate existing term, it is possible to create a **local* term within the **local** vocabulary. In this case describe your term carefully, add it to the database, and use it for content creation.
|
|
|
+ Creation of **local** terms is discouraged but sometimes necessary. When creating local terms, be careful in your description.
|
|
|
|
|
|
-Add the Vocabulary Term
|
|
|
-Once you've choosen a term to describe your content type, you should add the term to Tripal. If the term belongs to a controlled vocabulary with an file in OBO format then you can load all the terms of the vocabulary using Tripal's OBO Loader at Tripal → Data Loaders → Chado OBO Loader. See the Controlled Vocabularies section of the tutorial for more details. For adding a single term either from an existing vocabulary or a new local term, navigate to Tripal → Data Loaders → Chado Controlled Vocabularies and search to see if the vocabulary already exists. If it does you do not need to add the vocabulary. If it does not exist, click the Add Vocabulary link to add the vocabulary for your term. Then navigate to Tripal → Data Loaders → Chado CV Terms then click the Add Term link to add the term.
|
|
|
+How to Add a CV Term
|
|
|
+--------------------
|
|
|
+Loading From an OBO File
|
|
|
+^^^^^^^^^^^^^^^^^^^^^^^^
|
|
|
+Once you've choosen a term to describe your content type, you may need to add the term to Tripal if it is not already present. Many CVs use the `OBO file format <https://owlcollab.github.io/oboformat/doc/GO.format.obo-1_4.html>`_ to define their terms. If the term belongs to a controlled vocabulary with a file in OBO format then you can load all the terms of the vocabulary using Tripal's OBO Loader at **Tripal → Data Loaders → Chado Vocabularies → Chado OBO Loader**.
|
|
|
+
|
|
|
+Manually Adding a Term
|
|
|
+^^^^^^^^^^^^^^^^^^^^^^
|
|
|
+Alternatively, you can add terms one at a time. To add a single term either from an existing vocabulary or a new local term, navigate to **Tripal → Data Loaders → Chado Vocabularies → Manage Chado CVs** and search to see if the vocabulary already exists. If it does you do not need to add the vocabulary. If it does not exist, click the **Add Vocabulary** link to add the vocabulary for your term. Then navigate to **Tripal → Data Loaders → Chado Vocabularies → Mange Chado CV Terms** then click the **Add Term link** to add the term.
|
|
|
|
|
|
Create a Tripal Content Type
|
|
|
-Start by navigating to Structure → Tripal Content Types and then click on the Add Tripal Content Type link at the top. This will take you to a form that leads you through the process of creating a custom Tripal Content Type. First, enter for the name of the term you would like to use to describe your content in the Content Type autocomplete textbox (e.g. genetic_marker). This term cannot be changed later so make sure you put a lot of thought into it in the previous step. Then click Lookup Term. This should bring up a list of matching terms from which you can select the specific term you would like to use.
|
|
|
+----------------------------
|
|
|
|
|
|
+.. note::
|
|
|
|
|
|
+ Prior to creating a new content type you should understand the structure of Chado and how others store similar types of data.
|
|
|
|
|
|
-It will also bring up a section to specify which Chado tables you want the data for this content type stored in. Since genetic markers are sequence features, we would like to store them in the chado feature table. One you select the Chado table, the form will update with a number of leading questions to determine how to tell which records in a chado table belong to a given content type. Specifically, the following options are supported if the appropriate fields/tables are available:
|
|
|
+Creation of a new content type requires familiarity with Chado. This is because data records used by content types must be mapped to actual data and the data lives in Chado. Tripal's interface for creating content types allows you to provide the CV term for the type and then indicate where in Chado the data is/will be stored. Chado is a flexible relational database schema. Thus, it is possible for different sites to store data in different ways. It is best practice however to follow community standards when storing data. Therefore, please review the online documentation for Chado. If you are unclear how data for your content type should be stored in Chado please consider emailing the `Chado mailing list <http://gmod.org/wiki/GMOD_Mailing_Lists>`_ to ask for help or add a request for help on the Tripal issue queue.
|
|
|
|
|
|
-All records in the table belong to the current content type (e.g. organism)
|
|
|
-Records in the table where the type_id matches the content type term belong to the content type (e.g. genetic_marker)
|
|
|
-Records in the primary table that have a specific property stored in an associated *prop table (e.g. Genotyping Projects)
|
|
|
-Records in the primary table that have a specific annotated term in an associated *_cvterm table
|
|
|
-For genetic markers, we can simply use the type_id to differentiate which records in the feature table are genetic markers. Thus we
|
|
|
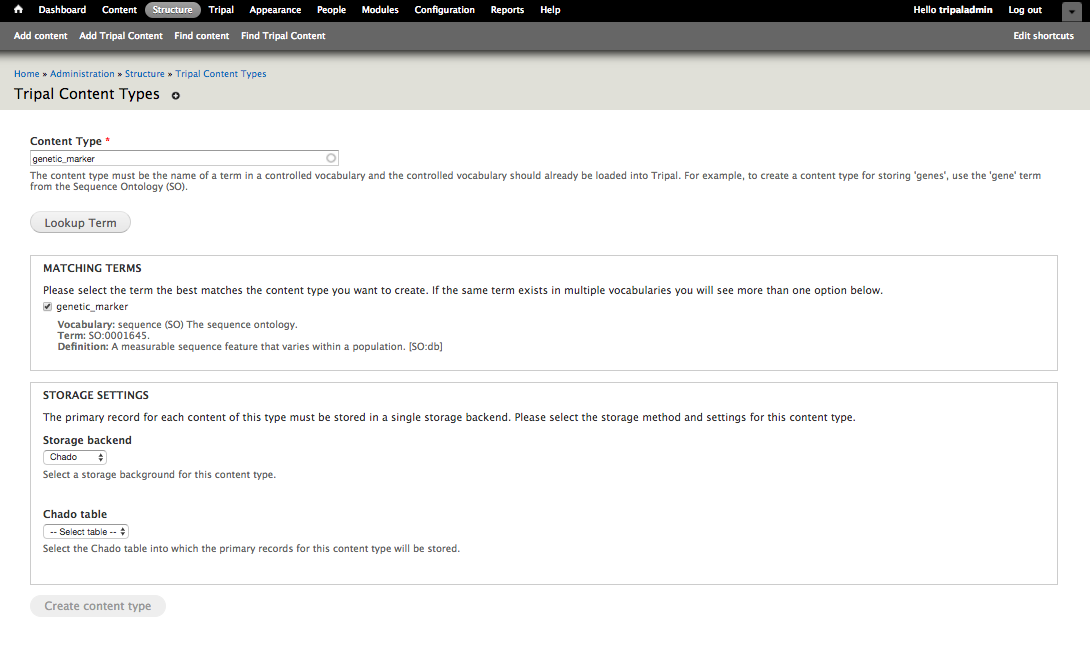
+To add a new content type, start by navigating to **Structure → Tripal Content Types** and click on the **Add Tripal Content Type** link at the top. This will take you to a web form that leads you through the process of creating a custom Tripal Content Type. First, enter for the name of the term you would like to use to describe your content in the Content Type autocomplete textbox (e.g. genetic_marker). Then, click **Lookup Term**. This should bring up a list of matching terms from which you can select the specific term you would like to use. Sometimes the same term exists in multiple vocabularies and you can select the proper one.
|
|
|
|
|
|
-Select "No" not all records in the feature table are genetic markers
|
|
|
-Type Column: type_id
|
|
|
-Then click Create Content Type to create a custom genetic marker content type.
|
|
|
+.. image:: creating_content.create1.png
|
|
|
|
|
|
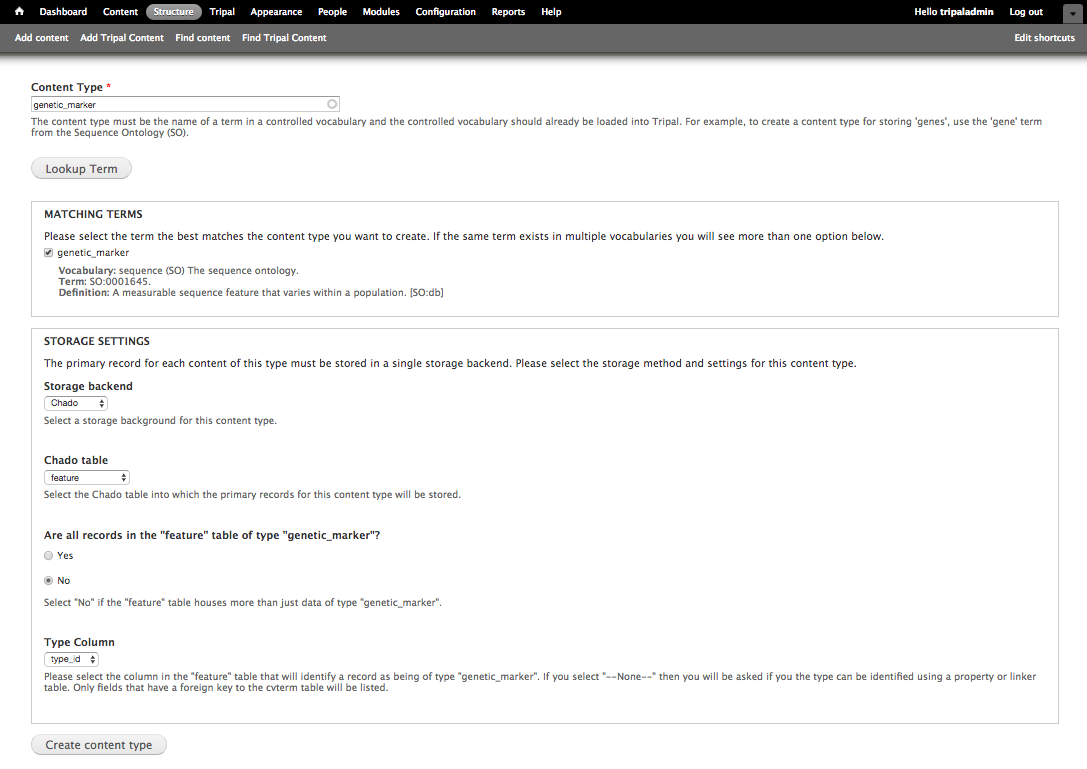
+During content type creation there is as a section to specify which Chado tables will store your data. Chado is typically structured with primary **base** tables (e.g. organism, feature, stock, project, etc) and a set of linker and property tables that contain ancillary data related to the base records. Here you must first choose the base table where primary records for your data type are stored. For our example, because genetic markers are sequence features, they are stored in the Chado feature table. Once you select the Chado table, the form will ask additional questions to determine exactly how records of the content type can be found. Specifically, the following options are supported if the appropriate fields/tables are available:
|
|
|
+
|
|
|
+1. All records in the **base** table belong to the content type (e.g. tables: organism, analysis, etc.)
|
|
|
+2. The **base** table has a **type_id** that stores the CV term and this differentiates the records. (e.g. tables: feature, stock, library, etc.).
|
|
|
+3. The records can be differentiated by way of a property table which contains a **type_id** column for the CV term. (e.g. tables: featureprop, stockprop, libraryprop, etc.)
|
|
|
+4. The records can be differentiated by way of a linking table that associates records in the **base** table with the CV term (e.g. tables: feature_cvterm, stock_cvterm, etc.)
|
|
|
+
|
|
|
+For our genetic marker example, we can use the Chado **feature** table and **type_id** column to differentiate which records in the feature table are genetic markers. Thus we
|
|
|
+
|
|
|
+- Select "No" not all records in the feature table are genetic markers
|
|
|
+- Type Column: type_id
|
|
|
+
|
|
|
+Then click Create Content Type to create a custom genetic marker content type.
|
|
|
|
|
|
+.. image:: creating_content.create2.png
|
|
|
|
|
|
-Once you create a custom content type, you can create pages of that type for your users either through the create tripal content user interface as we did for organisms or by loading your data into chado and then publishing it like we did for genes and mRNAs.
|
|
|
+Once the content type is created, you can create pages for site visitors. This will be described later in this User's Guide. In short, you can manually create new records through brand new web forms that are created automatically for your content type, or you can use a data loader to import your data directly to Chado, then **Publish** those records through the Tripal interface.
|