|
|
@@ -0,0 +1,531 @@
|
|
|
+Manual Field Creation
|
|
|
+======================
|
|
|
+
|
|
|
+To show how a TripalField works we will break down a class implementation section by section. Here we will use the **obi__organism** field that comes with Tripal and which extends the ChadoField class. The ChadoField class is almost identical to the TripalField class except that it provides a few extra settings for working with Chado tables. To create your own class you need to create a new class that implements the necessary functions.
|
|
|
+
|
|
|
+.. note::
|
|
|
+ Creation of your first field may not seem easy! The following document is a lot to think about and consider. Therefore, when you write your first field, don't try to do everything at once. Take it one piece at a time. The variables and functions described here are in order with the most critical components described first. Take it at an even pace.
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+Directory Structure for Fields
|
|
|
+~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+
|
|
|
+Before we create our class we must first create a proper directory structure. Tripal expects that all new Tripal field classes are located inside of a custom module in the following directory structure:
|
|
|
+
|
|
|
+.. code-block:: bash
|
|
|
+
|
|
|
+ /sites/all/modules/[your_module]/includes/TripalField/[field_name]/[field_name].inc
|
|
|
+ /sites/all/modules/[your_module]/includes/TripalField/[field_name]/[field_name]_widget.inc
|
|
|
+ /sites/all/modules/[your_module]/includes/TripalField/[field_name]/[field_name]_formatter.inc
|
|
|
+
|
|
|
+
|
|
|
+In the directories above the token [your_module] can be substituted with the name of your module and [field_name] is the name of your field. You can name your field whatever you like, but you must use this name consistently in other locations throughout the modules. Because all fields are defined by vocabulary terms, it is custom to name your fields with the vocabulary **short name** followed by two underscores followed by the **term name**, hence: obi__organism. Here the ChadoField implementation goes in the [field_name].inc file, the ChadoFieldWidget in the [field_name]_widget.inc file and the ChadoFieldFormatter in the [field_name]_formatter.inc. All new fields must implement all three classes. in the case of our obi__organism field the directory structure is as follows:
|
|
|
+
|
|
|
+.. code-block:: bash
|
|
|
+
|
|
|
+ /sites/all/modules/tripal/tripal_chado/includes/TripalField/obi__organism/obi__organism.inc
|
|
|
+ /sites/all/modules/tripal/tripal_chado/includes/TripalField/obi__organism/obi__organism_widget.inc
|
|
|
+ /sites/all/modules/tripal/tripal_chado/includes/TripalField/obi__organism/obi__organism_formatter.inc
|
|
|
+
|
|
|
+Anatomy of the ChadoField Class
|
|
|
+~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+The following describes a ChadoField class from top to bottom. The code for the obi__organism field is shown in order that it appears in the class with descriptions provided for the meaning of each piece of code. To write your own class, duplicate the variables and function and customize accordingly. First, let's look at the definition of the class. The following line defines the class and indicates that it extends the ChadoField class:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ class obi__organism extends ChadoField {
|
|
|
+
|
|
|
+.. note::
|
|
|
+
|
|
|
+ In the line above, the class is named obi__organism. This must be the same name as the directory in which the field is located. Otherwise, Tripal won't be able to find the field.
|
|
|
+
|
|
|
+Static Member Variables
|
|
|
+~~~~~~~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+Next, the TripalField/ChadoField class has a section of public static variables. These are variables that you can customize to describe your field to Tripal. Here you will provide the default label that appears for the field, and a description for the field:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ // The default label for this field.
|
|
|
+ public static $default_label = 'Organism';
|
|
|
+
|
|
|
+ // The default description for this field.
|
|
|
+ public static $description = 'The organism to which this resource is associated.';
|
|
|
+
|
|
|
+As described in the section titled Tripal Data Structures, fields that are attached to Bundles are "instances" of a field. Every field instance can be customized differently on each bundle. The following section of code allows your field to provide custom settings. Here we want to "hard-code" the term that defines this field using the $default_instance_settings variable:
|
|
|
+
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+ // Provide a list of instance specific settings. These can be access within
|
|
|
+ // the instanceSettingsForm. When the instanceSettingsForm is submitted
|
|
|
+ // then Drupal with automatically change these settings for the instance.
|
|
|
+ // It is recommended to put settings at the instance level whenever possible.
|
|
|
+ // If you override this variable in a child class be sure to replicate the
|
|
|
+ // term_name, term_vocab, term_accession and term_fixed keys as these are
|
|
|
+ // required for all TripalFields.
|
|
|
+ public static $default_instance_settings = array(
|
|
|
+ // The short name for the vocabulary (e.g. shcema, SO, GO, PATO, etc.).
|
|
|
+ 'term_vocabulary' => 'OBI',
|
|
|
+ // The name of the term.
|
|
|
+ 'term_name' => 'organism',
|
|
|
+ // The unique ID (i.e. accession) of the term.
|
|
|
+ 'term_accession' => '0100026',
|
|
|
+ // Set to TRUE if the site admin is allowed to change the term
|
|
|
+ // type. This will create form elements when editing the field instance
|
|
|
+ // to allow the site admin to change the term settings above.
|
|
|
+ 'term_fixed' => FALSE,
|
|
|
+ // The format for display of the organism.
|
|
|
+ 'field_display_string' => '<i>[organism.genus] [organism.species]</i>',
|
|
|
+ );
|
|
|
+
|
|
|
+Notice in the code above that the elements **term_vocabulary, term_name** and **term_accession** are used to define the vocabulary term that this field maps to. The term_fixed element allows the term to be changed by the site admin if desired. These elements are required of all TripalFields classes. You must always have these elements. However, the **field_display_string** is a variable unique to this obi__organism field! Because this field is displaying the organism we want to allow the site-admin to customize how the organism name is constructed and displayed. Therefore, the **field_display_string** creates this new setting for us. How this setting is used will be described later.
|
|
|
+
|
|
|
+As you may have noticed, a field requires a widget and a formatter. This is why there are three classes for every field. However, Drupal is flexible and allows fields to be edited or displayed by any number of widgets and formatters. By default, Tripal provides one widget class and one formatter class for every field. When you write a new field you will need to do the same and create a new ChadoFieldWidget and ChadoFieldFormatter class (or the corresponding non-Chado versions if you don't need Chado). The following variables in the class indicate what are the default widget and formatter classes (we have not yet created those, but we know their names!):
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+ // The default widget for this field.
|
|
|
+ public static $default_widget = 'obi__organism_widget';
|
|
|
+
|
|
|
+ // The default formatter for this field.
|
|
|
+ public static $default_formatter = 'obi__organism_formatter';
|
|
|
+
|
|
|
+Drupal allows new instances of fields to be attached to any Bundle. This is really useful for fields like the built in Image field that Drupal provides. It can be very handy to attache an instance of an Image field to any content type and viola! your content type now supports images. However, there are some fields that should never be added via the online Drupal interface. Our organism field is a good example. We probably don't want to allow end-users to add an organism field to a Person content type... In this case we will programmatically control which fields are attached to which Bundles. We'll show that later. But for now, let's set the no_ui variable to TRUE to prevent users from adding our new field to any Bundle.
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+ // A boolean specifying that users should not be allowed to create
|
|
|
+ // fields and instances of this field type through the UI. Such
|
|
|
+ // fields can only be created programmatically with field_create_field()
|
|
|
+ // and field_create_instance().
|
|
|
+ public static $no_ui = TRUE;
|
|
|
+
|
|
|
+
|
|
|
+Sometimes a field is meant to provide a visualization or some other functionality. An example of this might be a small search form or link to an analytical service. In these cases we want the field to show up on the web page but it should not appear anywhere else, such as in Tripal's web service that provides access to all content. We can set the no_data variable to TRUE and this will allow it to be seen on the site, but not anywhere else.
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+ // A boolean specifying that the field will not contain any data. This
|
|
|
+ // should exclude the field from web serivces or downloads. An example
|
|
|
+ // could be a quick search field that appears on the page that redirects
|
|
|
+ // the user but otherwise provides no data.
|
|
|
+
|
|
|
+
|
|
|
+ public static $no_data = FALSE;
|
|
|
+
|
|
|
+
|
|
|
+ .. note::
|
|
|
+ Be sure to only set this to TRUE when you are absolutely certain the contents would not be needed in web services. Tripal was designed so that what appears on the page will always appear in web services. Aside form the formatting we see on the website, the content should be the same.
|
|
|
+
|
|
|
+Finally, the last item in our Class variables is the **download_formatters**. Tripal provides an API that allows tools to group entities into data collections. Data collections are like "baskets" or "shopping carts". Entities that are in data collections can be downloaded into files. If your field is compatible with specific file downloaders you can specify those here. A file downloader is a special TripalFieldDownloader class that "speaks" certain file formats. Tripal, by default, provides the TripalTabDownloader (for tab-delimited files), the TripalCSVDownloader (for CSV files), a TripalNucFASTADownloader for creating nucleotide FASTA files and a TripalProteinFASTADownloader for protein FASTA files. If your field is compatible with any of these formatters you can specify them in the following array:
|
|
|
+
|
|
|
+.. .. code-block::
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ // Indicates the download formats for this field. The list must be the
|
|
|
+ // name of a child class of the TripalFieldDownloader.
|
|
|
+ public static $download_formatters = array(
|
|
|
+ 'TripalTabDownloader',
|
|
|
+ 'TripalCSVDownloader',
|
|
|
+ );
|
|
|
+
|
|
|
+
|
|
|
+If your field is compatible with the TripalTabDownloader, for example, your field will be included as a column in a tab delimited file where each row represents contents for a given entity.
|
|
|
+
|
|
|
+The load() function.
|
|
|
+~~~~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+The first function we want to implement in our class is the load() function. This function is responsible for querying the database and populating the field value. Data that is loaded into the field must be organized in two ways: 1) a value that is visible to the end-users, and 2) values that are visible to Chado for ensuing update/editing of the correct record in Chado when the field is edited. Our obi__organism field is designed to be used for multiple Bundles therefore the code in our load() function must be able to support any Chado table that has a foreign key relationship with the organism table.
|
|
|
+
|
|
|
+To get started, the load() function receives a single argument. The entity object:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ public function load($entity) {
|
|
|
+
|
|
|
+
|
|
|
+Because this is a ChadoField and the TripalChado module supports this field and maps entities to their "base" record on Chado, we get something extra... we get the record itself
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ $record = $entity->chado_record;
|
|
|
+
|
|
|
+Having the record helps tremendously. Our **obi__organism** field is meant to be attached to genomic feature content types (e.g. genes, mRNA, etc.), germplasm, etc. Therefore, the entity will be a record of one of those types. In the case of a genomic feature, these come from the **feature** table of Chado. In the case of a germplam, these records come from the **stock** table of Chado. Both of these records have an **organism_id** field which is a foreign key to the organism table where we find out details about the organism.
|
|
|
+
|
|
|
+Before we set the values for our field, we need a little bit more information. Remember that all field instances have settings? The Tripal Chado module also populates for us the name of the Chado table and the column that this field maps to. Our obi__organism field can be used for multiple Bundles. A gene bundle would map to the **feature** table of Chado and a germplasm Bundle would map to the **stock** table. We need to know what table and column this field is mapping to: We can get that from the instance object of the class and its settings:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ $settings = $this->instance['settings'];
|
|
|
+ $field_table = $this->instance['settings']['chado_table'];
|
|
|
+ $field_column = $this->instance['settings']['chado_column'];
|
|
|
+
|
|
|
+Next, we want to get this field name and its type. We obviously know our field name, it is obi__organism. However, we can get the name programmatically as well. Drupal maintains an "informational" array about our field. Inside of that field array we can find lots of interesting information such as our field name and its type (Bundle). We'll need this when we set our field value. But rather than hard-code it, let's grab it programmatically from the field name. It's best to grab it programmatically because there are cases where the field name could change:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ $field_name = $this->field['field_name'];
|
|
|
+ $field_type = $this->field['type'];
|
|
|
+
|
|
|
+
|
|
|
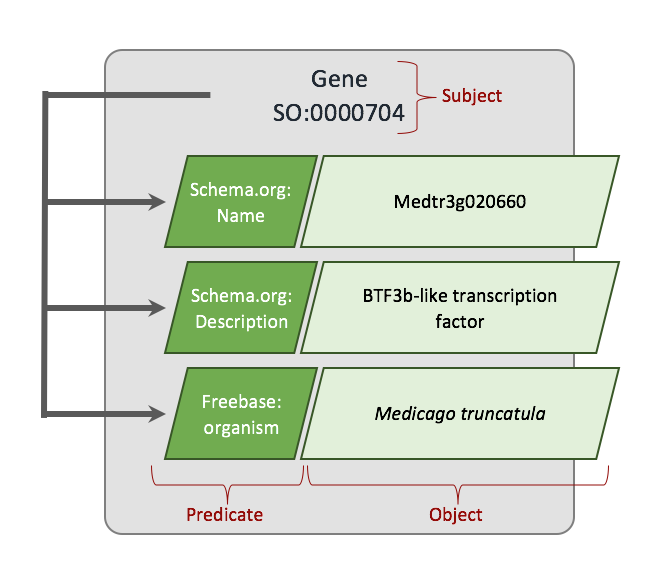
+Now, let's plan how we want our values to appear in our field. The organism record of Chado v1.3 has a genus, species, abbreviation, infraspecific name, infraspecific type, and a common name. We want these values exposed to the end user. But, wait... when we discussed fields in the Tripal Data Structures section we learned about a name field that provides names for entities. That field only has one value: the name. Our organism field has multiple values (i.e. genus, species, etc.). A field can provide more than just one value but values have to be qualified. We have to provide values in key/value pairs, and the keys must be controlled vocabulary terms. We must use controlled vocabulary terms because we want our field to be searchable by other Tripal sites. For example, the ontology term for the word 'genus' comes from the TAXRANK vocabulary. Fortunately, almost every column of every table in Chado has been mapped to a controlled vocabulary term so we don't need to go hunting for terms. We can use a Chado API function that Tripal provides for getting the ontology terms associated with every column table in Chado. The following code shows these functions retrieving the ontology terms for our values from the organism table:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ // Get the terms for each of the keys for the 'values' property.
|
|
|
+ $label_term = 'rdfs:label';
|
|
|
+ $genus_term = tripal_get_chado_semweb_term('organism', 'genus');
|
|
|
+ $species_term = tripal_get_chado_semweb_term('organism', 'species');
|
|
|
+ $infraspecific_name_term = tripal_get_chado_semweb_term('organism', 'infraspecific_name');
|
|
|
+ $infraspecific_type_term = tripal_get_chado_semweb_term('organism', 'type_id');
|
|
|
+
|
|
|
+
|
|
|
+Notice that for our organism fields we can easily get the ontology terms for them using the API function **tripal_get_chado_semweb_term**. You will also notice a **label_term** variable. Sometimes a user may want to see the full name of the organism and not pieces of it in various elements. Therefore, we will provide a label in our list of values that will concatenate the full organism name. This field is not in our organism table so we hard-code the term 'rdfs:lable' which is a term from the Resource Data Framework Schema vocabulary that defines a label.
|
|
|
+
|
|
|
+Next, let's initialize our field's value to be empty. When setting a field value we must do so in the entity object that got passed into our load function. The entity is an object and it stores values using the names of the fields. The following code sets an empty record for our field:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ // Set some defaults for the empty record.
|
|
|
+ $entity->{$field_name}['und'][0] = array(
|
|
|
+ 'value' => array(),
|
|
|
+ );
|
|
|
+
|
|
|
+
|
|
|
+Notice that our field has some sub elements. The first is 'und'. This element corresponds to the "language" of the text. Drupal supports mulitple spoken languages and wants to know the language of text we provide. For Tripal fields we always use 'und' meaning 'undefined'. The next element is the delta index number. Field have a cardinality, or in other words they can have multiple values. For every value we add we increment that index, always starting at zero. The last element is our 'value' element and it is here where we put our element. You may notice that our **delta** index is hard coded to 0. This is because an entity can only always have one organism that it is associated with. We will never have more than one.
|
|
|
+
|
|
|
+Now that we've got some preliminary values and we've initialized our value array we can start adding values! Before we do though, let's double check that we have a record. If we don't have a record for this entity, we can't get a value.
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ if ($record) {
|
|
|
+
|
|
|
+
|
|
|
+Now if we do have a record we need to get the value The first step is to actually get our organism record. For this we will find the record variable to be really handy. It already comes pre-populated with every Chado record that has a foreign-key relationship with our base record. So, in the case of a gene, the record is stored in the feature table which has an organism_id column which is a foreign key to the organism table. So, we know then that our record object has an organism_id property and we can get our organism from that. The only exception is the biomaterial table which uses a field named taxon_id:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ if ($field_table == 'biomaterial') {
|
|
|
+ $organism = $record->taxon_id;
|
|
|
+ }
|
|
|
+ else {
|
|
|
+ $organism = $record->organism_id;
|
|
|
+ }
|
|
|
+
|
|
|
+We can easily get all of the values we need from this organism object. We can now access the values for this organism using the Chado organism table column names (e.g. $organism->genus, $organism->species).
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $label = tripal_replace_chado_tokens($string, $organism);
|
|
|
+ $entity->{$field_name}['und'][0]['value'] = array(
|
|
|
+ $label_term => $label,
|
|
|
+ $genus_term => $organism->genus,
|
|
|
+ $species_term => $organism->species,
|
|
|
+ );
|
|
|
+ // The infraspecific fields were introduced in Chado v1.3.
|
|
|
+ if (property_exists($organism, 'infraspecific_name')) {
|
|
|
+ $entity->{$field_name}['und'][0]['value'][$infraspecific_type_term] = NULL;
|
|
|
+ $entity->{$field_name}['und'][0]['value'][$infraspecific_name_term] = $organism->infraspecific_name;
|
|
|
+ if ($organism->type_id) {
|
|
|
+ $entity->{$field_name}['und'][0]['value'][$infraspecific_type_term] = $organism->type_id->name;
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+In the code above we are populating our value array and we're using the controlled vocabulary terms we retrieved earlier as the keys.
|
|
|
+
|
|
|
+Okay, so, we have our values set. However, remember, our fields must support two types of values: 1) those for end users; and 2) those that allow us to save values in Chado if the field is edited. If you look at our value array above you will recognize that the entity to which this field is loading data for is for a feature or stock or library, etc. This field represents the organism for a record from one of those tables. If someone wants to edit the entity and change the organism then effectively we need to change the organism_id of that table. But in our values array we don't have the organism_id we only have data about the organism. How will Tripal know how to change the organism for an entity if edited? To do help Tripal out, we have to create special key/value pair to add to our values. These are values that are not meant to be seen by the end-user. The organism_id is a good example of such a value. To create these values we create a key with a special naming scheme: use "chado-" as a prefix, followed by the table name (e.g. feature), followed by two underscores and finally the column name (e.g. organism_id). The following code shows the creation of this value name:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ // Set the linker field appropriately.
|
|
|
+ if ($field_table == 'biomaterial') {
|
|
|
+ $linker_field = 'chado-biomaterial__taxon_id';
|
|
|
+ }
|
|
|
+ else {
|
|
|
+ $linker_field = 'chado-' . $field_table . '__organism_id';
|
|
|
+ }
|
|
|
+
|
|
|
+If our entity were of type "gene" then our **field_table** is feature. Therefore, our **linker_field** variable would be **chado-feature__organism_id**. Next, we need to add this to our value:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ $entity->{$field_name}['und'][0][$linker_field] = $organism->organism_id;
|
|
|
+
|
|
|
+Notice, though, that we did not add this value inside the 'value' key like we did above for our end-user, such as the following:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $entity->{$field_name}['und'][0]['value'])
|
|
|
+
|
|
|
+Instead, we put it in at the same level as 'value':
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $entity->{$field_name}['und'][0][$linker_field]
|
|
|
+
|
|
|
+We do this because anything in the 'value' element is intended for the end-user. Anything outside of the 'value' is meant for Tripal. Adding the organism ID to this field as a Tripal "hidden" value allows Tripal to recognize where these values really came from. When writing your own fields, you must include any values as "hidden" Tripal values that need to be written to the database table. A good way to remember if you a value should be visible to the end-user or hidden for Tripal is to ask yourself these questions:
|
|
|
+
|
|
|
+ 1. Does the user need this value? If yes, put it in the 'value' element.
|
|
|
+ 2. Does Tripal need the value when writing back to the Chado table? If yes, put it as a hidden element.
|
|
|
+ 3. Does the user need to see the value an will this same value need to be written to the table? If yes, then you have to put the value in both places.
|
|
|
+
|
|
|
+For our **obi__organism** field it is for entities with records in the **feature, stock, library**, etc. tables. Those tables only have an **organism_id** to represent the organism. So, that's the database column this field is supporting. We therefore, need to put that field as a hidden field, and all the others are just helpful to the user and don't get saved in the feature, stock or library tables. So, those go in the values array.
|
|
|
+
|
|
|
+Now, we're at a good stopping point with our field! We can close out our if($record) statement and the function:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+elementInfo() function
|
|
|
+~~~~~~~~~~~~~~~~~~~~~~
|
|
|
+The elementInfo() function is necessary to integrate your new field with Drupal Views and Tripal Web Services. Drupal needs to know what data elements your field provides and Tripal needs to know what vocabulary terms to use for each of the data elements. Related to vocabulary terms, all fields are assigned an ontology term for the field itself. Every field has to have an one. But when a field provides more than just a single data value it must also provide vocabulary terms for any sub elements as well. Our obi__organism field provides the genus, species, etc. sub elements and, therefore, we need to describe these to Drupal and Tripal. The elementInfo() function from the obi_organism field is as follows:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ /**
|
|
|
+ * @see TripalField::elementInfo()
|
|
|
+ */
|
|
|
+ public function elementInfo() {
|
|
|
+ $field_term = $this->getFieldTermID();
|
|
|
+
|
|
|
+ $genus_term = chado_get_semweb_term('organism', 'genus');
|
|
|
+ $species_term = chado_get_semweb_term('organism', 'species');
|
|
|
+ $infraspecific_name_term = chado_get_semweb_term('organism', 'infraspecific_name');
|
|
|
+ $infraspecific_type_term = chado_get_semweb_term('organism', 'type_id');
|
|
|
+
|
|
|
+ return array(
|
|
|
+ $field_term => array(
|
|
|
+ 'operations' => array('eq', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:complexType',
|
|
|
+ 'elements' => array(
|
|
|
+ 'rdfs:label' => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'scientific_name',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'readonly' => TRUE,
|
|
|
+ 'required' => FALSE,
|
|
|
+ ),
|
|
|
+ $genus_term => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'genus',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'required' => TRUE,
|
|
|
+ ),
|
|
|
+ $species_term => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'species',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'required' => TRUE,
|
|
|
+ ),
|
|
|
+ $infraspecific_name_term => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'infraspecies',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'required' => FALSE,
|
|
|
+ ),
|
|
|
+ $infraspecific_type_term => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'infraspecific_type',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:integer',
|
|
|
+ 'required' => FALSE,
|
|
|
+ ),
|
|
|
+ 'entity' => array(
|
|
|
+ 'searchable' => FALSE,
|
|
|
+ ),
|
|
|
+ ),
|
|
|
+ ),
|
|
|
+ );
|
|
|
+ }
|
|
|
+
|
|
|
+
|
|
|
+The code above generates and returns an associative array that provides metadata about the field and its elements. The array is structured such that the first-level key is the term for the field. Details about the field are at the second-level and all sub elements are contained in a 'elements' key. In the following code the terms for the field and sub elements are retrieved using TripalField class functions and Tripal API calls:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $field_term = $this->getFieldTermID();
|
|
|
+
|
|
|
+ $genus_term = chado_get_semweb_term('organism', 'genus');
|
|
|
+ $species_term = chado_get_semweb_term('organism', 'species');
|
|
|
+ $infraspecific_name_term = chado_get_semweb_term('organism', 'infraspecific_name');
|
|
|
+ $infraspecific_type_term = chado_get_semweb_term('organism', 'type_id');
|
|
|
+
|
|
|
+ return array( $field_term => array(
|
|
|
+ 'operations' => array('eq', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:complexType',
|
|
|
+ 'elements' => array(
|
|
|
+
|
|
|
+Notice the value for $field_term variable was easily obtained by calling the $this->getFieldTermID function and all of the terms for the elements were obtained using the chado_get_semweb_term function which maps table columns in the Chado database schema to ontology terms. The operations key indicates which search filter operations are supported for the field as a whole. For this example these include 'eq' (for equals), 'contains' and 'starts' (for starts with). The field is sortable and searchable so those values are set to TRUE. Later, we weill learn how to implement the sorting, searching and filtering that the field will support. For now we know we want them so we set the values accordingly. Additionally, the field allows updating so 'readonly' is set to FALSE. By convention, the 'type' of a field follows the XML data types for simple types (https://www.w3schools.com/xml/schema_simple.asp) and Complex types (https://www.w3schools.com/xml/schema_complex.asp) that have multiple elements. Because our obi__organism field has subelements and is not a single value, the field type is 'xs:complexType'.
|
|
|
+
|
|
|
+The array keys just mentioned fully describe our field to Drupal and Tripal. Next we will define the sub elements in the same way, and these go in the 'elements' key. First, we will describe the label. Our obi__oranganism field provides a handly label element that concatenates the genus, species and infraspecific name into one simple string. Therefore, we need to describe this element in the same way we described the field itself. In the code below that the key is set to 'rdfs:label' (which is the controlled vocabulary term for a label) and that the child keys are the same as for the field.
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+
|
|
|
+ 'elements' => array(
|
|
|
+ 'rdfs:label' => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'scientific_name',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'readonly' => TRUE,
|
|
|
+ 'required' => FALSE,
|
|
|
+ ),
|
|
|
+
|
|
|
+Notice that our field will allow searching, provides a variety of search filter options, is sortable and defines the type as 'xs:string'. The remaing elements follow the same pattern. As another example, here is the genus element:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $genus_term => array(
|
|
|
+ 'searchable' => TRUE,
|
|
|
+ 'name' => 'genus',
|
|
|
+ 'operations' => array('eq', 'ne', 'contains', 'starts'),
|
|
|
+ 'sortable' => TRUE,
|
|
|
+ 'readonly' => FALSE,
|
|
|
+ 'type' => 'xs:string',
|
|
|
+ 'required' => TRUE,
|
|
|
+ ),
|
|
|
+
|
|
|
+The major difference in the code above is that the term is provided by the variable $genus_term.
|
|
|
+
|
|
|
+Finally, our obi__organism field provides an 'entity' element that provides information for a published organism entity. We do not provide any filtering, searching or sorting of those values. So the final element appears as:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ 'entity' => array(
|
|
|
+ 'searchable' => FALSE,
|
|
|
+ ),
|
|
|
+
|
|
|
+In summary, you will always want to describe your field and every element of your field in the array returned by the elementInfo function. However, you do not need to provide sorting, filtering or querying for every element. If your field is read-only and simply provides values you should still describe these elements but you would set the meta data keys appropriately for the behavior of your field. Also, you only need to describe elements in the values array returned by your load function. Remember, there may be other key/value pairs (such as those used to help coordinate inserts/updates into Chado) but those do not need to be described here because they are never seen by the end-user.
|
|
|
+
|
|
|
+query() function
|
|
|
+~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+
|
|
|
+As described above in the elementInfo function section, some fields and elements of fields are searchable. if the elementInfo array indicates that the field is searchable and has operations (i.e. filters) then we must provide a way for those queries to occur. This is where the query() function is needed. The following is example code from the query function of our obi__organism field:
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ public function query($query, $condition) {
|
|
|
+ $alias = $this->field['field_name'];
|
|
|
+ $operator = $condition['operator'];
|
|
|
+
|
|
|
+ $field_term_id = $this->getFieldTermID();
|
|
|
+ $genus_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'genus');
|
|
|
+ $species_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'species');
|
|
|
+ $infraspecific_name_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'infraspecific_name');
|
|
|
+ $infraspecific_type_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'type_id');
|
|
|
+
|
|
|
+ // Join to the organism table for this field.
|
|
|
+ $this->queryJoinOnce($query, 'organism', $alias, "base.organism_id = $alias.organism_id");
|
|
|
+
|
|
|
+ // If the column is the field name then we're during a search on the full
|
|
|
+ // scientific name.
|
|
|
+ if ($condition['column'] == $field_term_id or
|
|
|
+ $condition['column'] == $field_term_id . ',rdfs:label') {
|
|
|
+ if (chado_get_version() <= 1.3) {
|
|
|
+ $query->where("CONCAT($alias.genus, ' ', $alias.species) $operator :full_name", array(':full_name' => $condition['value']));
|
|
|
+ }
|
|
|
+ else {
|
|
|
+ $this->queryJoinOnce($query, 'cvterm', $alias . '_cvterm', 'base.infraspecific_type = ' . $alias . '_cvterm.type_id', 'LEFT OUTER');
|
|
|
+ $query->where("CONCAT($alias.genus, ' ', $alias.species, ' ', " . $alias . "'_cvterm.name', ' ', $alias.infraspecific_name) $operator :full_name", array(':full_name' => $condition['value']));
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ // If the column is a subfield.
|
|
|
+ if ($condition['column'] == $species_term) {
|
|
|
+ $query->condition("$alias.species", $condition['value'], $operator);
|
|
|
+ }
|
|
|
+ if ($condition['column'] == $genus_term) {
|
|
|
+ $query->condition("$alias.genus", $condition['value'], $operator);
|
|
|
+ }
|
|
|
+
|
|
|
+ if ($condition['column'] == $infraspecific_name_term) {
|
|
|
+ $query->condition("$alias.infraspecific_name", $condition['value'], $operator);
|
|
|
+ }
|
|
|
+
|
|
|
+ if ($condition['column'] == $infraspecific_type_term) {
|
|
|
+ $this->queryJoinOnce($query, 'cvterm', 'CVT', "base.type_id = CVT.cvterm_id");
|
|
|
+ $query->condition("CVT.name", $condition['value'], $operator);
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+The code above is how the field tells Drupal and Tripal how to find and filter the records that this field corresponds to. First, we retreive the field alias and operators:and as with the load and elementInfo functions we get the controlled vocabulary terms for our field and field elements:
|
|
|
+
|
|
|
+
|
|
|
+.. code-block:: php
|
|
|
+
|
|
|
+ <?php
|
|
|
+
|
|
|
+ $alias = $this->field['field_name'];
|
|
|
+ $operator = $condition['operator'];
|
|
|
+
|
|
|
+ $field_term_id = $this->getFieldTermID();
|
|
|
+ $genus_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'genus');
|
|
|
+ $species_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'species');
|
|
|
+ $infraspecific_name_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'infraspecific_name');
|
|
|
+ $infraspecific_type_term = $field_term_id . ',' . chado_get_semweb_term('organism', 'type_id');
|
|
|
+
|
|
|
+Next, our knowledge of Chado is needed. We know that our obi__organism field will load data from the organism table. Therefore, our search must occur there.
|