+ 1
- 0
docs/dev_guide.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/dev_guide/exporting_field_settings.1.assign_term.png
二进制
docs/dev_guide/exporting_field_settings.2.png
二进制
docs/dev_guide/exporting_field_settings.3.png
二进制
docs/dev_guide/exporting_field_settings.4.png
二进制
docs/dev_guide/exporting_field_settings.5.png
二进制
docs/dev_guide/exporting_field_settings.6.png
二进制
docs/dev_guide/exporting_field_settings.7.png
+ 121
- 0
docs/dev_guide/exporting_field_settings.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
docs/user_guide.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 2
- 1
docs/user_guide/content_types.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 3
- 1
docs/user_guide/content_types/creating_content.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/user_guide/content_types/field_permissions.1.cross_ref_GA.png
二进制
docs/user_guide/content_types/field_permissions.2.crossref_permissions.png

+ 56
- 0
docs/user_guide/content_types/field_permissions.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 0
docs/user_guide/example_genomics.rst
|
||
|
||
|
||
|
||
|
||
+ 14
- 0
docs/user_guide/example_genomics/func_annots.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 175
- 0
docs/user_guide/example_genomics/func_annots/blast.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/user_guide/example_genomics/func_annots/blast1.png
二进制
docs/user_guide/example_genomics/func_annots/blast2.png
二进制
docs/user_guide/example_genomics/func_annots/blast3.png
二进制
docs/user_guide/example_genomics/func_annots/blast4.png
二进制
docs/user_guide/example_genomics/func_annots/blast5.png
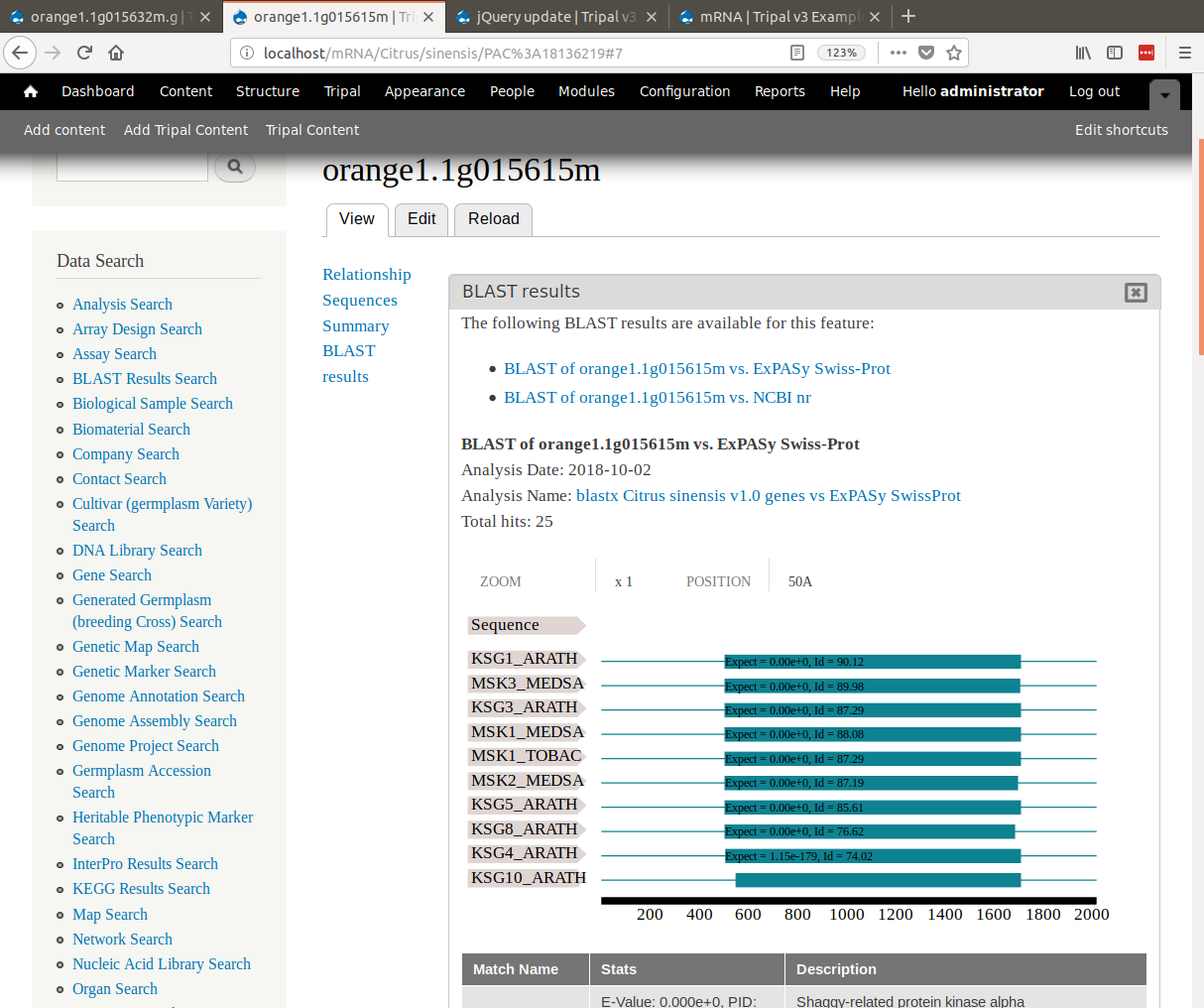
二进制
docs/user_guide/example_genomics/func_annots/blast6.png
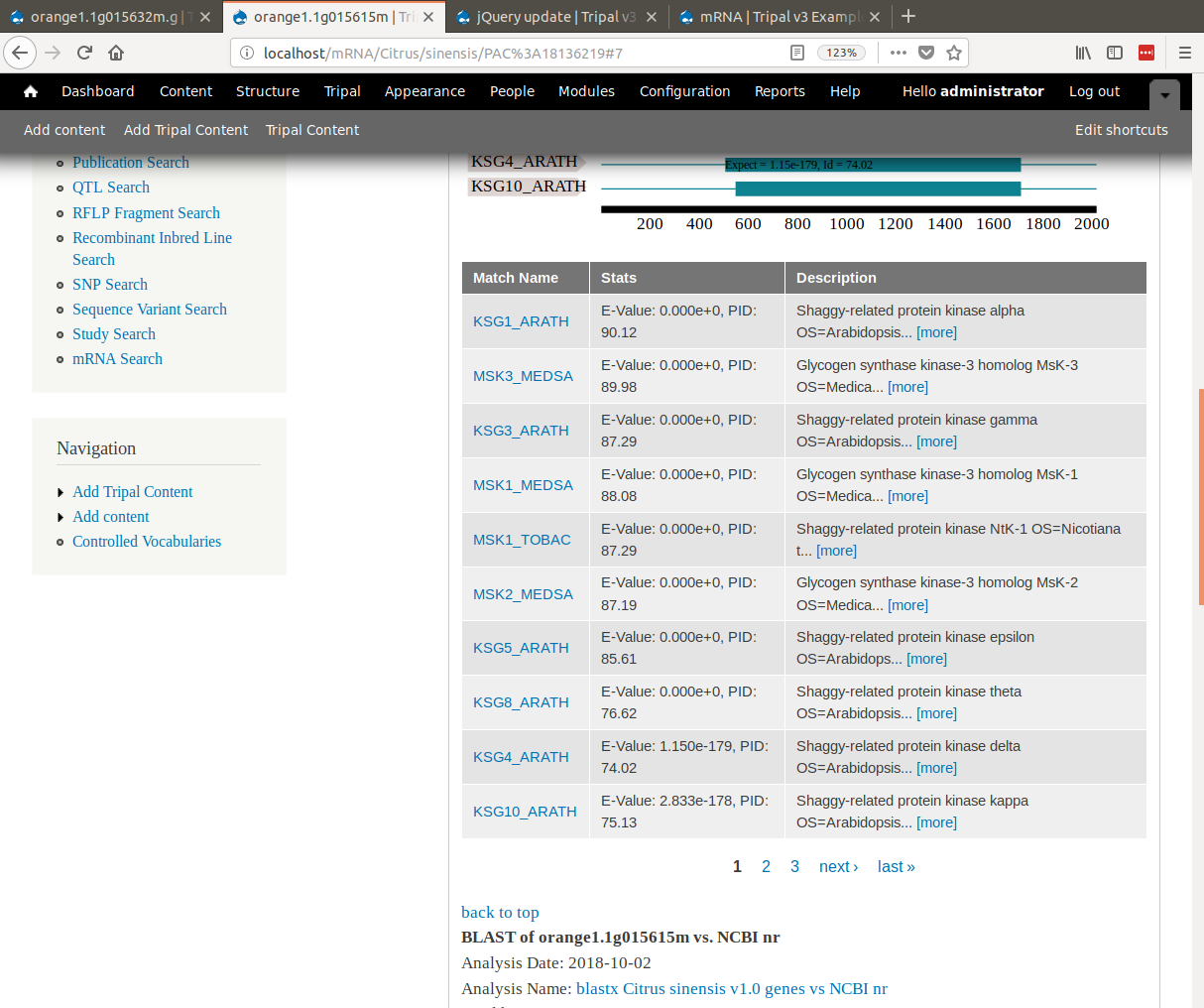
+ 2
- 0
docs/user_guide/example_genomics/func_annots/go.rst
|
||
|
||
|
||
+ 80
- 0
docs/user_guide/example_genomics/func_annots/interpro.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/user_guide/example_genomics/func_annots/interpro1.png
二进制
docs/user_guide/example_genomics/func_annots/interpro2.png
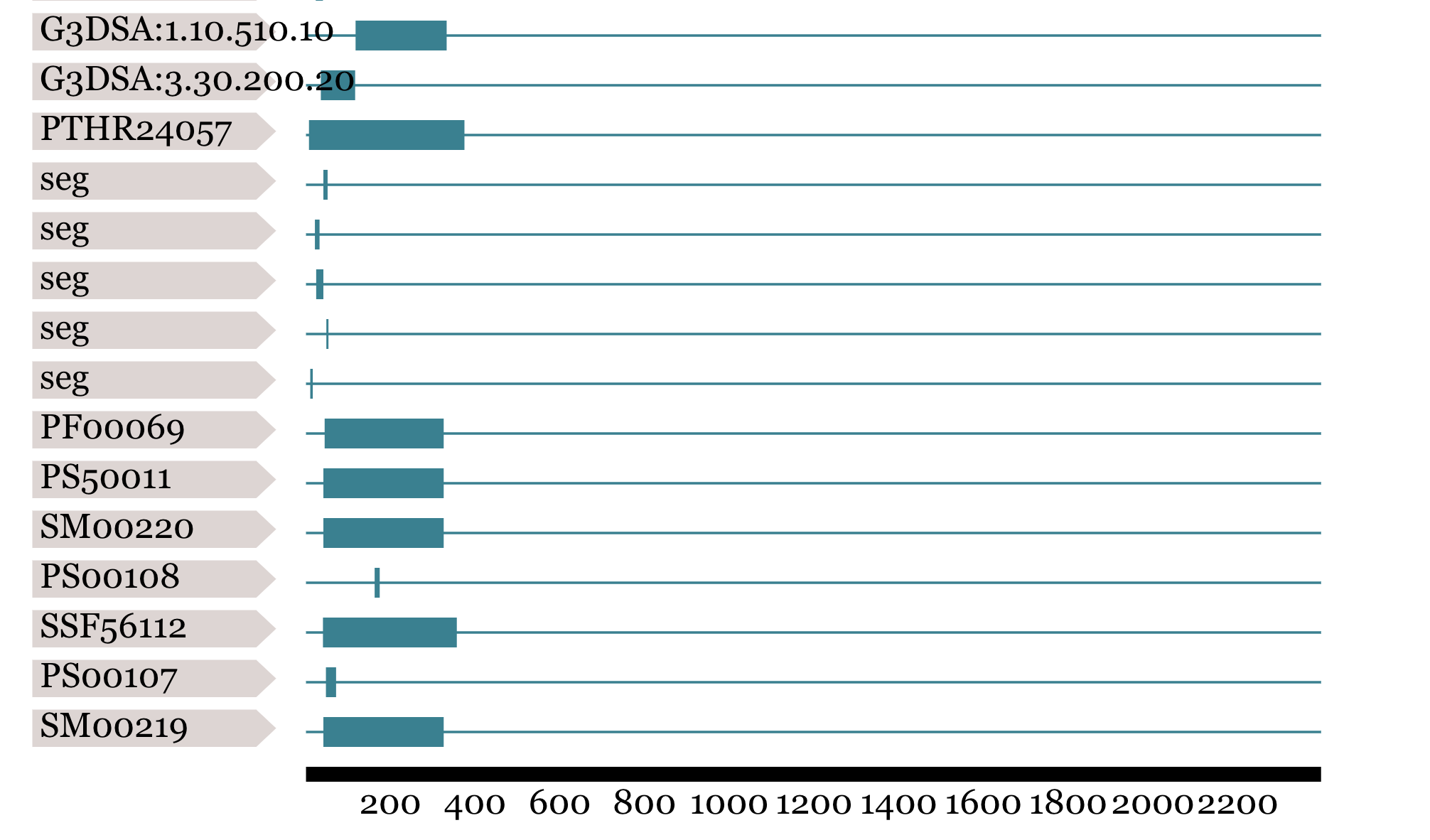
+ 2
- 0
docs/user_guide/example_genomics/func_annots/kegg.rst
|
||
|
||
|
||
+ 73
- 0
docs/user_guide/example_genomics/func_annots/setup.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/user_guide/example_genomics/func_annots/setup1.png
二进制
docs/user_guide/example_genomics/func_annots/setup2.png
+ 1
- 1
docs/user_guide/example_genomics/organisms.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
二进制
docs/user_guide/searching/default_pages.1.png
+ 9
- 0
docs/user_guide/searching/default_pages.rst
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_analysis/tripal_analysis.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_contact/tripal_contact.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_core/tripal_core.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_cv/tripal_cv.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_db/tripal_db.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_feature/tripal_feature.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_featuremap/tripal_featuremap.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_genetic/tripal_genetic.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_library/tripal_library.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_natural_diversity/tripal_natural_diversity.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_organism/tripal_organism.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_phenotype/tripal_phenotype.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_phylogeny/tripal_phylogeny.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_project/tripal_project.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_pub/tripal_pub.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_stock/tripal_stock.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
legacy/tripal_views/tripal_views.info
|
||
|
||
|
||
|
||
|
||
|
||
+ 4
- 1
tripal/includes/TripalFieldQuery.inc
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
tripal/tripal.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
tripal_bulk_loader/tripal_bulk_loader.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
tripal_chado/tripal_chado.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
tripal_chado_views/tripal_chado_views.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 2
- 12
tripal_chado_views/tripal_chado_views.module
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
tripal_ds/tripal_ds.info
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||