|
|
@@ -3,41 +3,37 @@ Module Setup
|
|
|
.. note::
|
|
|
|
|
|
Remember you must set the ``$DRUPAL_HOME`` environment variable if you want to cut-and-paste the commands below. See :doc:`../../install_tripal/drupal_home`
|
|
|
-
|
|
|
-
|
|
|
-For this example we will be load functional data for our gene. To do this we will use the Blast, KEGG, and InterPro extension modules. However, these extension modules are not part of the "core" Tripal package but are available as separate extensions. Anyone may create extensions for Tripal. These extensions are useful for genomic data and therefore are included in this tutorial.
|
|
|
+
|
|
|
+
|
|
|
+For this example we will be load functional data for our gene. To do this we will use the Blast, KEGG, and InterPro extension modules. However, these extension modules are not part of the "core" Tripal package but are available as separate extensions. Anyone may create extensions for Tripal. These extensions are useful for genomic data and therefore are included in this tutorial.
|
|
|
|
|
|
To download these modules:
|
|
|
|
|
|
::
|
|
|
-
|
|
|
- cd $DRUPAL_HOME
|
|
|
+
|
|
|
+ cd $DRUPAL_HOME
|
|
|
drush pm-download tripal_analysis_blast
|
|
|
- drush pm-download tripal_analysis_kegg
|
|
|
drush pm-download tripal_analysis_interpro
|
|
|
|
|
|
Now, enable these extension modules:
|
|
|
|
|
|
::
|
|
|
-
|
|
|
+
|
|
|
drush pm-enable tripal_analysis_blast
|
|
|
drush pm-enable tripal_analysis_interpro
|
|
|
- drush pm-enable tripal_analysis_kegg
|
|
|
|
|
|
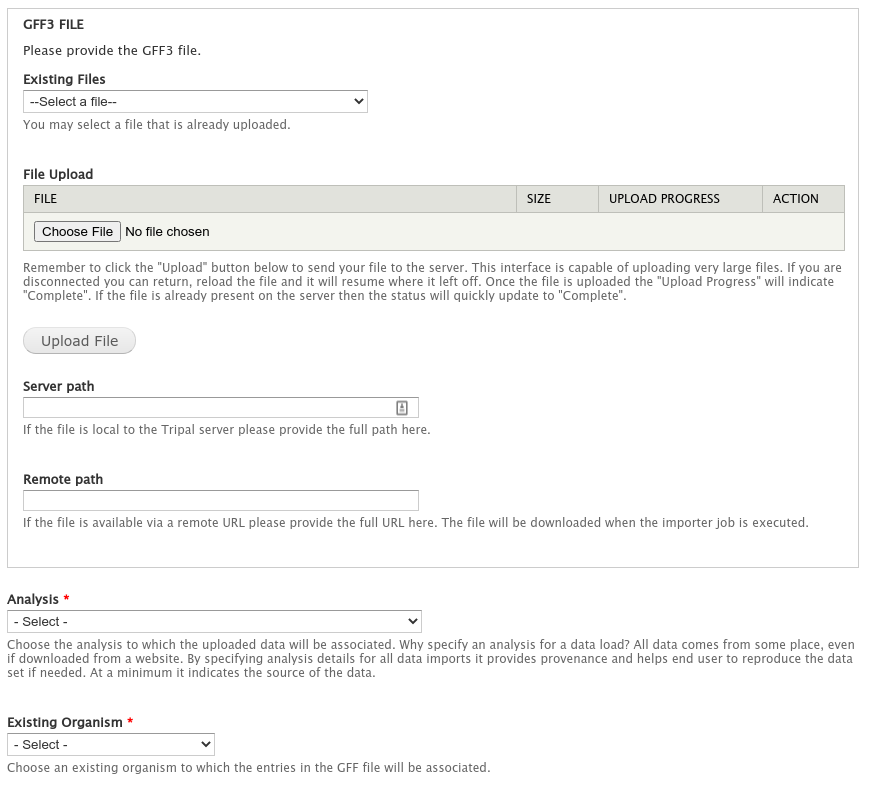
For this example, we will use the following files which are available for downloading:
|
|
|
|
|
|
- `Citrus_sinensis-orange1.1g015632m.g.iprscan.xml <http://www.gmod.org/mediawiki/images/0/0c/Citrus_sinensis-orange1.1g015632m.g.iprscan.xml>`_
|
|
|
-- `Citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz <http://www.gmod.org/mediawiki/images/1/13/Citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz>`_
|
|
|
- `Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out <http://www.gmod.org/mediawiki/images/e/e8/Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out>`_
|
|
|
- `Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out <http://www.gmod.org/mediawiki/images/2/24/Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out>`_
|
|
|
|
|
|
Download these files to the ```$DRUPAL_HOME/sites/default/files``` directory. To do so quickly run these commands:
|
|
|
|
|
|
::
|
|
|
-
|
|
|
+
|
|
|
cd $DRUPAL_HOME/sites/default/files
|
|
|
wget http://www.gmod.org/mediawiki/images/0/0c/Citrus_sinensis-orange1.1g015632m.g.iprscan.xml
|
|
|
- wget http://www.gmod.org/mediawiki/images/1/13/Citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz
|
|
|
wget http://www.gmod.org/mediawiki/images/e/e8/Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out
|
|
|
wget http://www.gmod.org/mediawiki/images/2/24/Blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out
|
|
|
|
|
|
@@ -45,23 +41,22 @@ Each of these modules provides new fields for both the **gene** and **mRNA** con
|
|
|
|
|
|
.. image:: setup1.png
|
|
|
|
|
|
-Next, we need to position the new field. Using the skills you learned in the :doc:`../../content_types/configuring_page_display` Create three new **Tripal Panes** named:
|
|
|
+Next, we need to position the new field. Using the skills you learned in the :doc:`../../content_types/configuring_page_display` Create two new **Tripal Panes** named:
|
|
|
|
|
|
- Blast Results
|
|
|
- Protein Domains
|
|
|
-- KEGG Pathways
|
|
|
|
|
|
Be sure to:
|
|
|
|
|
|
-- Place the three new fields into each pane respectively
|
|
|
-- Move the Panes out of the **disabled** section.
|
|
|
+- Place the new fields into each pane respectively
|
|
|
+- Move the Panes out of the **disabled** section.
|
|
|
- Set the label for each field to **Hidden**.
|
|
|
|
|
|
The following shows an example of this layout:
|
|
|
|
|
|
.. image:: setup2.png
|
|
|
|
|
|
-The fields are now ready for display once data is added!
|
|
|
+The fields are now ready for display once data is added!
|
|
|
|
|
|
.. note::
|
|
|
|
|
|
@@ -70,4 +65,3 @@ The fields are now ready for display once data is added!
|
|
|
.. note::
|
|
|
|
|
|
Anytime you install a Tripal v3 extension module you should check for new fields, and then place those fields in the layout. Extension modules often will not do this for you because they do not assume you want these new fields.
|
|
|
-
|