|
|
@@ -38,7 +38,7 @@ This file has three columns: NCBI taxonomy ID, genus and species:
|
|
|
To use the bulk loader you must be familiar with the Chado database schema and have an idea for where data should be stored. It is best practice to consult the GMOD website or consult the Chado community (via the `gmod-schema mailing list <https://lists.sourceforge.net/lists/listinfo/gmod-schema>`_) when deciding how to store data. For this example, we want to add the species to Chado, and we want to associate the NCBI taxonomy ID with these organisms. The first step, therefore, is to decide where in Chado these data should go. In Chado, organisms are stored in the **organism** table. This table has the following fields:
|
|
|
|
|
|
.. csv-table:: Chado organism table
|
|
|
- header: "Name", "Type", "Description"
|
|
|
+ :header: "Name", "Type", "Description"
|
|
|
|
|
|
"organism_id", "serial", "PRIMARY KEY"
|
|
|
"abbreviation", "character varying(255)",
|
|
|
@@ -46,43 +46,40 @@ To use the bulk loader you must be familiar with the Chado database schema and h
|
|
|
"species", "character varying(255)", "UNIQUE#1 NOT NULL A type of organism is always uniquely identified by genus and species. When mapping from the NCBI taxonomy names.dmp file, this column must be used where it is present, as the common_name column is not always unique (e.g. environmental samples). If a particular strain or subspecies is to be represented, this is appended onto the species name. Follows standard NCBI taxonomy pattern."
|
|
|
"common_name", "character varying(255)"
|
|
|
"comment", "text"
|
|
|
+
|
|
|
+
|
|
|
We can therefore store the second and third columns of the tab-delimited input file in the **genus** and **species** columns of the organism table.
|
|
|
|
|
|
In order to store a database external reference (such as for the NCBI Taxonomy ID) we need to use the following tables: **db**, **dbxref**, and **organism_dbxref**. The **db** table will house the entry for the NCBI Taxonomy; the **dbxref** table will house the entry for the taxonomy ID; and the **organism_dbxref** table will link the taxonomy ID stored in the **dbxref** table with the organism housed in the **organism** table. For reference, the fields of these tables are as follows:
|
|
|
|
|
|
|
|
|
|
|
|
-db Structure
|
|
|
-F-Key Name Type Description
|
|
|
- db_id serial PRIMARY KEY
|
|
|
- name character varying(255) UNIQUE NOT NULL
|
|
|
- description character varying(255)
|
|
|
- urlprefix character varying(255)
|
|
|
- url character varying(255)
|
|
|
+.. csv-table:: chado.db structure
|
|
|
+ :header: "Name", "Type", "Description"
|
|
|
|
|
|
+ "db_id", "serial", "PRIMARY KEY"
|
|
|
+ "name", character varying(255), "UNIQUE NOT NULL"
|
|
|
+ "description", "character varying(255)", ""
|
|
|
+ "urlprefix", "character varying(255)"
|
|
|
+ "url", "character varying(255)"
|
|
|
|
|
|
-dbxref Structure
|
|
|
-F-Key Name Type Description
|
|
|
- dbxref_id serial PRIMARY KEY
|
|
|
-db
|
|
|
|
|
|
-db_id integer UNIQUE#1 NOT NULL
|
|
|
- accession character varying(255) UNIQUE#1 NOT NULL
|
|
|
-The local part of the identifier. Guaranteed by the db authority to be unique for that db.
|
|
|
+.. csv-table:: chado.dbxref structure
|
|
|
+ :header: "Name", "Type", "Description"
|
|
|
|
|
|
- version character varying(255) UNIQUE#1 NOT NULL DEFAULT ''::character varying
|
|
|
- description text
|
|
|
+ "dbxref_id", "serial", "PRIMARY KEY"
|
|
|
+ "db_id", "integer", "Foreign Key db. UNIQUE#1 NOT NULL"
|
|
|
+ "accession", "character varying(255)", "UNIQUE#1 NOT NULL. The local part of the identifier. Guaranteed by the db authority to be unique for that db."
|
|
|
+ "version", "character varying(255)", "UNIQUE#1 NOT NULL DEFAULT ''"
|
|
|
+ "description", "text"
|
|
|
|
|
|
|
|
|
-organism_dbxref Structure
|
|
|
-FK Name Type Description
|
|
|
- organism_dbxref_id serial PRIMARY KEY
|
|
|
-organism
|
|
|
+.. csv-table:: chado.organism_dbxref structure
|
|
|
+ :header: "Name", "Type", "Description"
|
|
|
|
|
|
-organism_id integer UNIQUE#1 NOT NULL
|
|
|
-dbxref
|
|
|
-
|
|
|
-dbxref_id integer UNIQUE#1 NOT NULL
|
|
|
+ "organism_dbxref_id", "serial", "PRIMARY KEY"
|
|
|
+ "organism_id", "integer", "Foreign key organism. UNIQUE#1 NOT NULL"
|
|
|
+ "dbxref_id", "integer", "Foreign key dbxref. UNIQUE#1 NOT NULL"
|
|
|
|
|
|
|
|
|
For our bulk loader template, we will therefore need to insert values into the **organism**, **db**, **dbxref** and **organism_dbxref** tables. In our input file we have the genus and species and taxonomy ID so we can import these with a bulk loader template. However, we do not have information that will go into the db table (e.g. "NCBI Taxonomy"). This is not a problem as the bulk loader can use existing data to help with import. We simply need to use the "NCBI Taxonomy" database that is currently in the Chado instance of Tripal v3.
|
|
|
@@ -146,13 +143,13 @@ To this point, we have built the loader such that it can load two of the three c
|
|
|
* Unique Record Name: NCBI Taxonomy DB
|
|
|
* Record Type/Action: SELECT ONCE: Select the record only once for each constant set.
|
|
|
* For the field:
|
|
|
- * Type of field: Constant
|
|
|
- * Human-readable Title for Field: DB name
|
|
|
- * Chado table: db
|
|
|
- * Chado field/column: name
|
|
|
+ * Type of field: Constant
|
|
|
+ * Human-readable Title for Field: DB name
|
|
|
+ * Chado table: db
|
|
|
+ * Chado field/column: name
|
|
|
* Within the Constant section:
|
|
|
- * Constant Value: NCBITaxon
|
|
|
- * Check "Ensure the value is in the table"
|
|
|
+ * Constant Value: NCBITaxon
|
|
|
+ * Check "Ensure the value is in the table"
|
|
|
|
|
|
|
|
|
Here we use a field type of **Constant** rather than **Data**. This is because we are providing the value to be used in the record rather than using a value from the input file. The value we are providing is "NCBI Taxonomy" which is the name of the database we added previously. The goal is to match the name "NCBI Taxonomy" with an entry in the **db** table. Click the **Save Changes** button.
|
|
|
@@ -161,18 +158,20 @@ We now see a second record on the **Edit Template** page. However, the mode for
|
|
|
|
|
|
Now that we have a record that selects the **db_id** we can now create the **dbxref** record. For the **dbxref** record there is a unique constraint that requires the **accession**, **db_id** and **version**. The version record has a default value so we only need to create two fields for this new record: the db_id and the accession. We will use the **db_id** from the **NCBI Taxonomy DB** record and the accession is the first column of the input file. First, we will add the **db_id** record. Click the **New Record/Field** button and set the following:
|
|
|
|
|
|
-For the record:
|
|
|
-Record: New Record
|
|
|
-Unique Record Name: Taxonomy ID
|
|
|
-Record Type/Action: INSERT: insert the record
|
|
|
-For the field:
|
|
|
-Type of field: Record referral
|
|
|
-Human-readable Title for Field: NCBI Taxonomy DB ID
|
|
|
-Chado table: dbxref
|
|
|
-Chado Field/Column: db_id
|
|
|
-In the Record Referral Section:
|
|
|
-Record to refer to: NCBI Taxonomy DB
|
|
|
-Field to refer to: db_id
|
|
|
+* For the record:
|
|
|
+ * Record: New Record
|
|
|
+ * Unique Record Name: Taxonomy ID
|
|
|
+ * Record Type/Action: INSERT: insert the record
|
|
|
+* For the field:
|
|
|
+ * Type of field: Record referral
|
|
|
+ * Human-readable Title for Field: NCBI Taxonomy DB ID
|
|
|
+ * Chado table: dbxref
|
|
|
+ * Chado Field/Column: db_id
|
|
|
+* In the Record Referral Section:
|
|
|
+ * Record to refer to: NCBI Taxonomy DB
|
|
|
+ * Field to refer to: db_id
|
|
|
+
|
|
|
+
|
|
|
Click the Save Changes button. The Edit Template page appears.
|
|
|
|
|
|
|
|
|
@@ -182,126 +181,147 @@ Click the Save Changes button. The Edit Template page appears.
|
|
|
|
|
|
Again, we need to edit the record to make the loader more fault tolerant. Click the Edit link to the left of the Taxonomy ID record. Select the following:
|
|
|
|
|
|
-Insert
|
|
|
-Select if duplicate
|
|
|
+* Insert
|
|
|
+* Select if duplicate
|
|
|
+
|
|
|
To complete this record, we need to add the accession field. Click the Add field link to the left of the Taxonomy ID record name. Provide the following values:
|
|
|
|
|
|
-For the field:
|
|
|
-Type of Field: Data
|
|
|
-Human-readable Title for Field: Accession
|
|
|
-Chado table: dbxref
|
|
|
-Chado field/column: accession
|
|
|
-In the Data File Column section:
|
|
|
-Column: 1
|
|
|
-At this state, we should have three records: Organism, NCBI Taxonomy DB, and Taxonomy ID. We can now add the final record that will insert a record into organism_dbxref table. Create this new record with the following details:
|
|
|
-
|
|
|
-Create Record with first field:
|
|
|
-
|
|
|
-For the record:
|
|
|
-Record: New Record
|
|
|
-Unique Record Name: Taxonomy/Organism Linker
|
|
|
-Check: Insert: insert the record
|
|
|
-For the field:
|
|
|
-Type of Field: Record Referral
|
|
|
-Human-readable Title for Field: Accession Ref
|
|
|
-Chado table: organism_dbxref
|
|
|
-Chado field/column: dbxref_id
|
|
|
-In the Record Referral section:
|
|
|
-Record to refer to: Taxonomy ID
|
|
|
-Field to refer to: dbxref_id
|
|
|
+* For the field:
|
|
|
+ * Type of Field: Data
|
|
|
+ * Human-readable Title for Field: Accession
|
|
|
+ * Chado table: dbxref
|
|
|
+ * Chado field/column: accession
|
|
|
+* In the Data File Column section:
|
|
|
+ * Column: 1
|
|
|
+
|
|
|
+At this state, we should have three records: Organism, NCBI Taxonomy DB, and Taxonomy ID. We can now add the final record that will insert a record into the **organism_dbxref** table. Create this new record with the following details:
|
|
|
+
|
|
|
+
|
|
|
+* For the record:
|
|
|
+ * Record: New Record
|
|
|
+ * Unique Record Name: Taxonomy/Organism Linker
|
|
|
+ * Check: Insert: insert the record
|
|
|
+* For the field:
|
|
|
+ * Type of Field: Record Referral
|
|
|
+ * Human-readable Title for Field: Accession Ref
|
|
|
+ * Chado table: organism_dbxref
|
|
|
+ * Chado field/column: dbxref_id
|
|
|
+* In the Record Referral section:
|
|
|
+ * Record to refer to: Taxonomy ID
|
|
|
+ * Field to refer to: dbxref_id
|
|
|
+
|
|
|
Create the second field:
|
|
|
|
|
|
-For the field:
|
|
|
-Type of Field: Record Referral
|
|
|
-Human-readable Title for Field: Organism ID
|
|
|
-Chado table: organism_dbxref
|
|
|
-Chado field/column: organism_id
|
|
|
-In the Record Referral section:
|
|
|
-Record to refer to: Organism
|
|
|
-Field to refer to: organism_id
|
|
|
+* For the field:
|
|
|
+ * Type of Field: Record Referral
|
|
|
+ * Human-readable Title for Field: Organism ID
|
|
|
+ * Chado table: organism_dbxref
|
|
|
+ * Chado field/column: organism_id
|
|
|
+* In the Record Referral section:
|
|
|
+ * Record to refer to: Organism
|
|
|
+ * Field to refer to: organism_id
|
|
|
+
|
|
|
After saving the field. Edit the record and set the following:
|
|
|
|
|
|
-Change the record mode to: insert or select if duplicate
|
|
|
-We are now done! We have created a bulk loader template that reads in a file with three columns containing an NCBI taxonomy ID, a genus and species. The loader places the genus and species in the organism table, adds the NCBI Taxonomy ID to the dbxref table, links it to the NCBI Taxonomy entry in the db table, and then adds an entry to the organism_dbxref table that links the organism to the NCBI taxonomy Id. The following screen shots show how the template should appear:
|
|
|
+* Change the record mode to: insert or select if duplicate
|
|
|
+
|
|
|
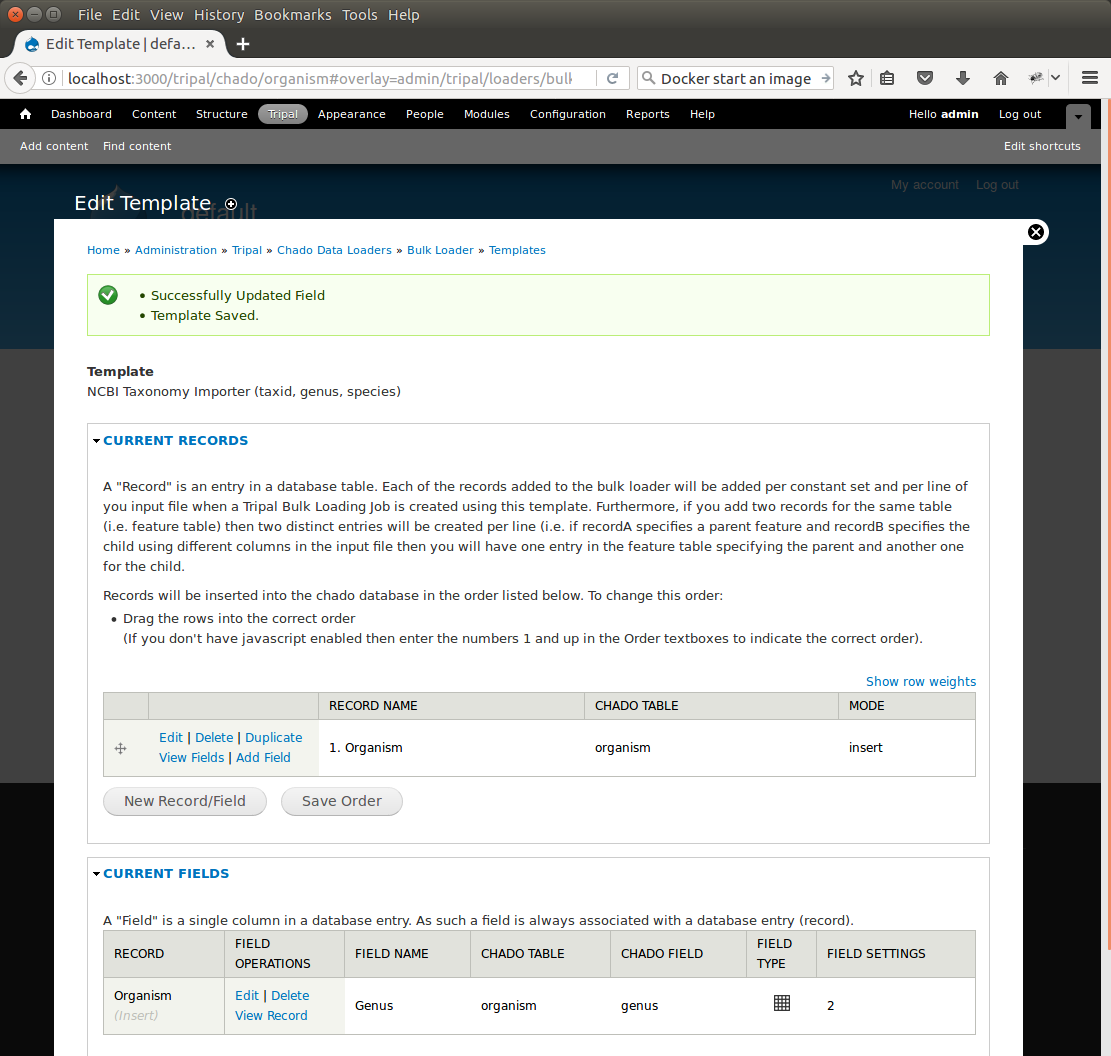
+We are now done! We have created a bulk loader template that reads in a file with three columns containing an NCBI taxonomy ID, a genus and species. The loader places the genus and species in the **organism** table, adds the NCBI Taxonomy ID to the **dbxref** table, links it to the NCBI Taxonomy entry in the db table, and then adds an entry to the **organism_dbxref** table that links the organism to the NCBI taxonomy Id. The following screen shots show how the template should appear:
|
|
|
|
|
|
|
|
|
.. image:: ./bulk_loader.7.png
|
|
|
|
|
|
|
|
|
-To save the template, click the Save Template link at the bottom of the page.
|
|
|
+To save the template, click the **Save Template** link at the bottom of the page.
|
|
|
|
|
|
Creating a Bulk Loader Job (importing a file)
|
|
|
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
|
|
|
|
|
|
-Now that we have created a bulk loader template we can use it to import a file. We will import the Fragaria.txt file downloaded previously. To import a file using a bulk loader template, click the Add Content link in the administrative menu and click the Bulk Loading Job. A bulk loading job is required each time we want to load a file. Below is a screen shot of the page used for creating a bulk loading job.
|
|
|
+Now that we have created a bulk loader template we can use it to import a file. We will import the **Fragaria**.txt file downloaded previously. To import a file using a bulk loader template, click the **Add Content** link in the administrative menu and click the **Bulk Loading Job**. A bulk loading job is required each time we want to load a file. Below is a screen shot of the page used for creating a bulk loading job.
|
|
|
|
|
|
|
|
|
|
|
|
Provide the following values:
|
|
|
|
|
|
-Job Name: Import of Fragaria species
|
|
|
-Template: NCBI Taxonomy Importer (taxid, genus species).
|
|
|
-Data File: /var/www/html/sites/default/files/Fragaria_0.txt
|
|
|
-Keep track of inserted IDs: No
|
|
|
-File has a header: No
|
|
|
-Click Save the page then appears as follows:
|
|
|
+* Job Name: Import of Fragaria species
|
|
|
+* Template: NCBI Taxonomy Importer (taxid, genus species).
|
|
|
+* Data File: /var/www/html/sites/default/files/Fragaria_0.txt
|
|
|
+* Keep track of inserted IDs: No
|
|
|
+* File has a header: No
|
|
|
+
|
|
|
+Click **Save**. The page then appears as follows:
|
|
|
|
|
|
|
|
|
.. image:: ./bulk_loader.8.png
|
|
|
|
|
|
|
|
|
-You can see details about constants that are used by the template and the where the fields from the input file will be stored by clicking the Data Fields tab in the table of contents on the left sidebar.
|
|
|
+You can see details about constants that are used by the template and the where the fields from the input file will be stored by clicking the **Data Fields** tab in the table of contents on the left sidebar.
|
|
|
|
|
|
|
|
|
.. image:: ./bulk_loader.9.png
|
|
|
|
|
|
|
|
|
-Now that we have created a job, we can submit it for execution by clicking the Submit Job button. This adds a job to the Tripal Jobs systems and we can launc the job as we have previously in this tutorial:
|
|
|
+Now that we have created a job, we can submit it for execution by clicking the **Submit Job** button. This adds a job to the Tripal Jobs systems and we can launc the job as we have previously in this tutorial:
|
|
|
+
|
|
|
+.. code-block:: shell
|
|
|
+
|
|
|
+ cd /var/www
|
|
|
+ drush trp-run-jobs --username=admin --root=/var/www/html
|
|
|
|
|
|
-cd /var/www
|
|
|
-drush trp-run-jobs --username=admin --root=/var/www/html
|
|
|
After execution of the job you should see similar output to the terminal window:
|
|
|
|
|
|
-Tripal Job Launcher
|
|
|
-Running as user 'admin'
|
|
|
--------------------
|
|
|
-There are 1 jobs queued.
|
|
|
-Calling: tripal_bulk_loader_load_data(2, 7)
|
|
|
-Template: NCBI Taxonomy Importer (taxid, genus, species) (1)
|
|
|
-File: /var/www/html/sites/default/files/Fragaria_0.txt (46 lines)
|
|
|
-
|
|
|
-Preparing to load...
|
|
|
-Loading...
|
|
|
- Preparing to load the current constant set...
|
|
|
- Open File...
|
|
|
- Start Transaction...
|
|
|
- Defer Constraints...
|
|
|
- Acquiring Table Locks...
|
|
|
- ROW EXCLUSIVE for organism
|
|
|
- ROW EXCLUSIVE for dbxref
|
|
|
- ROW EXCLUSIVE for organism_dbxref
|
|
|
- Loading the current constant set...
|
|
|
-Progress:
|
|
|
-[|||||||||||||||||||||||||||||||||||||||||||||||||||] 100.00%. (46 of 46) Memory: 33962080
|
|
|
-
|
|
|
-Our Fragaira species should now be loaded, and we return to the Tripal site to see them. Click on the Organisms link in the Search Data menu. In the search form that appears, type "Fragaria" in the Genus text box and click the Filter button. We should see the list of newly added Fragaria species.
|
|
|
+.. code-block:: shell
|
|
|
+
|
|
|
+
|
|
|
+ Tripal Job Launcher
|
|
|
+ Running as user 'admin'
|
|
|
+ -------------------
|
|
|
+ There are 1 jobs queued.
|
|
|
+ Calling: tripal_bulk_loader_load_data(2, 7)
|
|
|
+ Template: NCBI Taxonomy Importer (taxid, genus, species) (1)
|
|
|
+ File: /var/www/html/sites/default/files/Fragaria_0.txt (46 lines)
|
|
|
+
|
|
|
+ Preparing to load...
|
|
|
+ Loading...
|
|
|
+ Preparing to load the current constant set...
|
|
|
+ Open File...
|
|
|
+ Start Transaction...
|
|
|
+ Defer Constraints...
|
|
|
+ Acquiring Table Locks...
|
|
|
+ ROW EXCLUSIVE for organism
|
|
|
+ ROW EXCLUSIVE for dbxref
|
|
|
+ ROW EXCLUSIVE for organism_dbxref
|
|
|
+ Loading the current constant set...
|
|
|
+ Progress:
|
|
|
+ [|||||||||||||||||||||||||||||||||||||||||||||||||||] 100.00%. (46 of 46) Memory: 33962080
|
|
|
+
|
|
|
+Our *Fragaira* species should now be loaded, and we return to the Tripal site to see them. Click on the **Organisms** link in the **Search Data** menu. In the search form that appears, type "Fragaria" in the **Genus** text box and click the **Filter** button. We should see the list of newly added *Fragaria* species.
|
|
|
|
|
|
.. image:: ./bulk_loader.10.png
|
|
|
|
|
|
|
|
|
+Before the organisms will have Tripal pages, the Chado records need to be **Published**. You can publish them by navigating to ``admin -> Tripal Content -> Publish Tripal Content``. Select the **organism** table from the dropdown and run the job.
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+.. note::
|
|
|
|
|
|
-These Fragaria species are not synced and do not have pages which is why there are no links for them in the search results. To check if the NCBI taxonomy IDs are properly associated, we can sync all or some of them. We can sync organisms as per the instructions earlier in this Tutorial, but in short, navigate to Tripal → Chado Modules → Organisms, click the Sync tab and select the species you would like to sync, or select none and click the Sync Organisms button to sync them all. Execute the sync job. Once complete, return to the search form, find a Fragaria species that has been synced and view it's page. You should see a Cross References link in the left table of contents. If you click that link you should see the NCBI Taxonomy ID with a link to the page:
|
|
|
+ In Tripal 2, records were synced by naviating to ``Tripal → Chado Modules → Organisms``.
|
|
|
|
|
|
|
|
|
+
|
|
|
+Once complete, return to the search form, find a *Fragaria* species that has been published and view its page. You should see a Cross References link in the left table of contents. If you click that link you should see the NCBI Taxonomy ID with a link to the page:
|
|
|
+
|
|
|
.. image:: ./bulk_loader.11.png
|
|
|
|
|
|
|
|
|
Sharing Your Templates with Others
|
|
|
-Now that our template for loading organisms with NCBI Taxonomy IDs is completed we can share our template loader with anyone else that has a Tripal-based site. To do this we simply export the template in text format, place it in a text file or directly in an email and send to a collaborator for import into their site. To do this, navigate to Tripal → Chado Data Loaders → Buik Loader and click the Tempalate tab at the top. Here we find a table of all the tempaltes we have created. We should see our template named NCBI Taxonom Importer (taxid, genus, species). In the far right colum is a link to export that template. Licking that link will redirect you to a page where the template is provided in a serialized PHP array.
|
|
|
+~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
|
|
|
+
|
|
|
+Now that our template for loading organisms with NCBI Taxonomy IDs is completed we can share our template loader with anyone else that has a Tripal-based site. To do this we simply export the template in text format, place it in a text file or directly in an email and send to a collaborator for import into their site. To do this, navigate to ``Tripal → Chado Data Loaders → Buik Loader`` and click the **Tempalate** tab at the top. Here we find a table of all the tempaltes we have created. We should see our template named **NCBI Taxonomy Importer** (taxid, genus, species). In the far right colum is a link to export that template. Licking that link will redirect you to a page where the template is provided in a serialized PHP array.
|
|
|
|
|
|
.. image:: ./bulk_loader.12.png
|
|
|
|
|
|
|
|
|
-Simply cut-and-paste all of the text in the Export field and send it to a collaborator.
|
|
|
+Simply cut-and-paste all of the text in the **Export** field and send it to a collaborator.
|
|
|
|
|
|
-To import a template that may have been created by someone else, navigate to Tripal → Chado Data Loaders → Buik Loader and click the Tempalate tab. A link titled Import Template appears above the table of existing importers. The page that appears when that link is clicked will allow you to import any template shared with you.
|
|
|
+To import a template that may have been created by someone else, navigate to ``Tripal → Chado Data Loaders → Buik Loader`` and click the **Tempalate** tab. A link titled Import Template appears above the table of existing importers. The page that appears when that link is clicked will allow you to import any template shared with you.
|