|
|
@@ -191,7 +191,7 @@ Now the scaffold sequence and mRNA sequences are loaded!
|
|
|
It is not required to load the mRNA sequences as those can be derived from their alignments with the scaffold sequence. However, in Chado the **feature** table has a **residues** column. Therefore, it is best practice to load the sequence when possible.
|
|
|
|
|
|
Creating Gene Pages
|
|
|
-----------------------
|
|
|
+-------------------
|
|
|
Now that we've loaded our feature data, we must publish them. This is different than when we manually created our Organism and Analysis pages. Using the GFF and FASTA loaders we imported our data into Chado, but currently there are no published pages for this data that we loaded. To publish these genomic features, navigating to Structure → Tripal Content Types and click the link titled Publish Chado Content. The following page appears:
|
|
|
|
|
|
.. image:: genomes_genes.3.png
|
|
|
@@ -252,3 +252,29 @@ Next find an mRNA page to view. Remember when we loaded our FASTA file for mRNA
|
|
|
- We associated the Phytozome accession with the features using a regular expression when importing the FASTA file.
|
|
|
|
|
|
All data that appears on the page is derived from the GFF file and the FASTA files we loaded.
|
|
|
+
|
|
|
+
|
|
|
+Customizing Transcripts on Gene Pages
|
|
|
+-------------------------------------
|
|
|
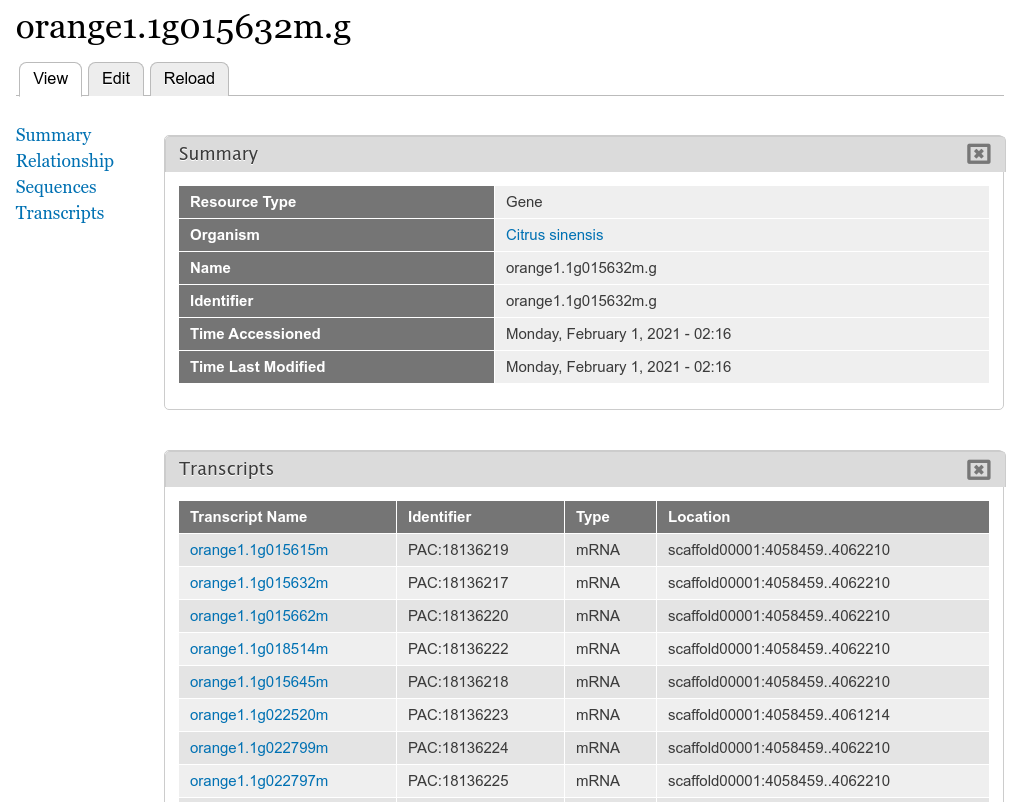
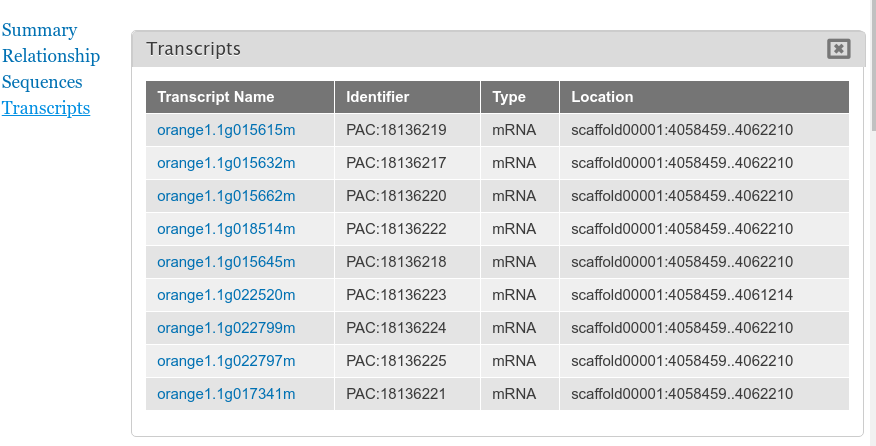
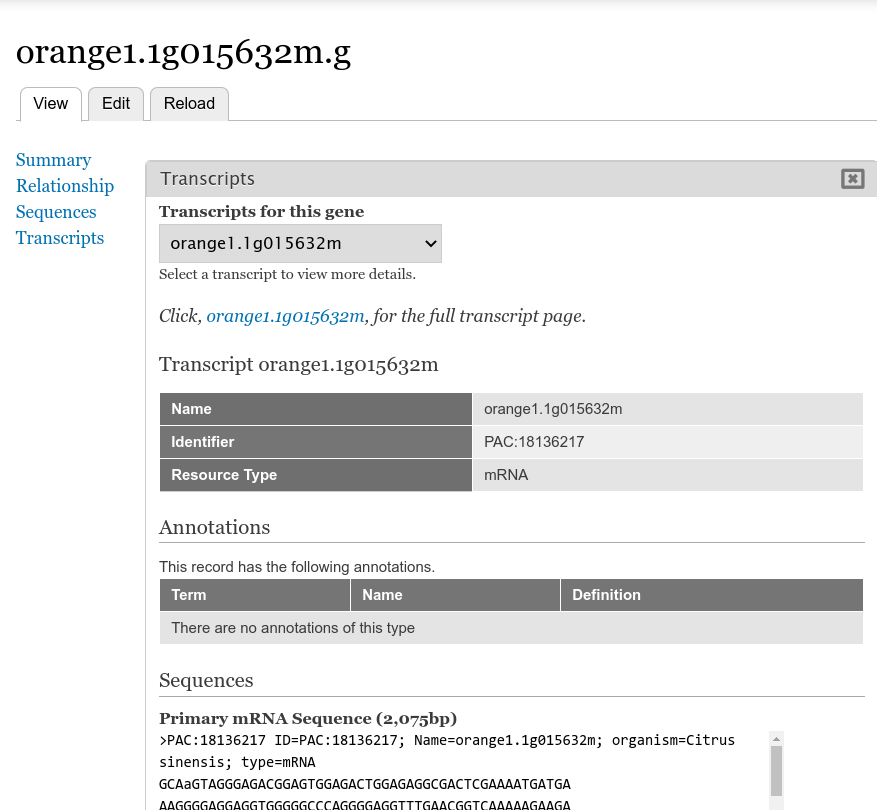
+By default the gene pages provided by Tripal will have a link in the sidebar table of contents named **Transcripts** and when clicked a table appears that lists all of the transcripts (or mRNA) that belong to the gene. The user can click to view more information about each published transcript.
|
|
|
+
|
|
|
+.. image:: genomes_genes.6.png
|
|
|
+
|
|
|
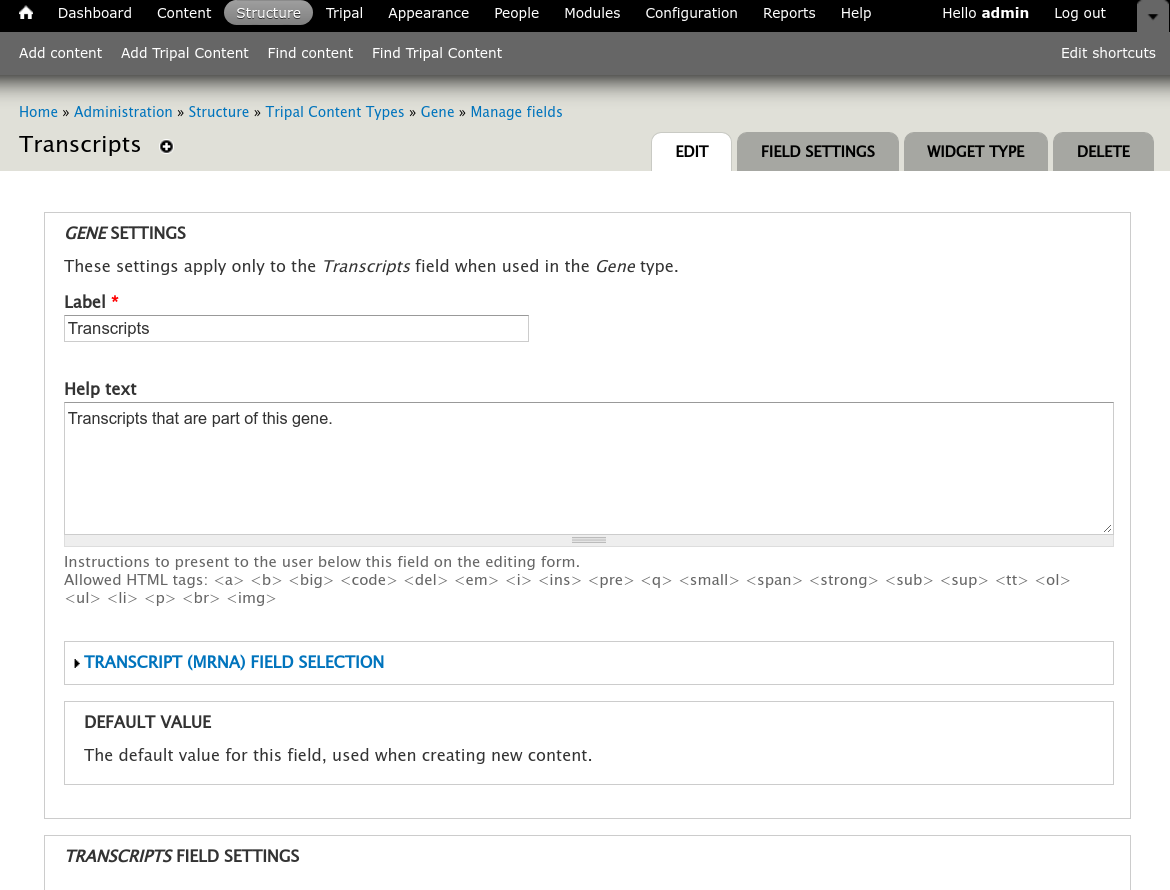
+Sometimes however, more than just a listing of transcripts is desired on a gene page. You can customize the information that is presented about each transcript by navigating to the gene content type at **Structure → Tripal Content Types** and clicking **mange fields** in the **Gene** row. This page allows you to customize the way fields are displayed on the gene page. Scroll down the page to the **Transcript** row and click the **edit** button. The following page should appear.
|
|
|
+
|
|
|
+.. image:: genomes_genes.7.png
|
|
|
+
|
|
|
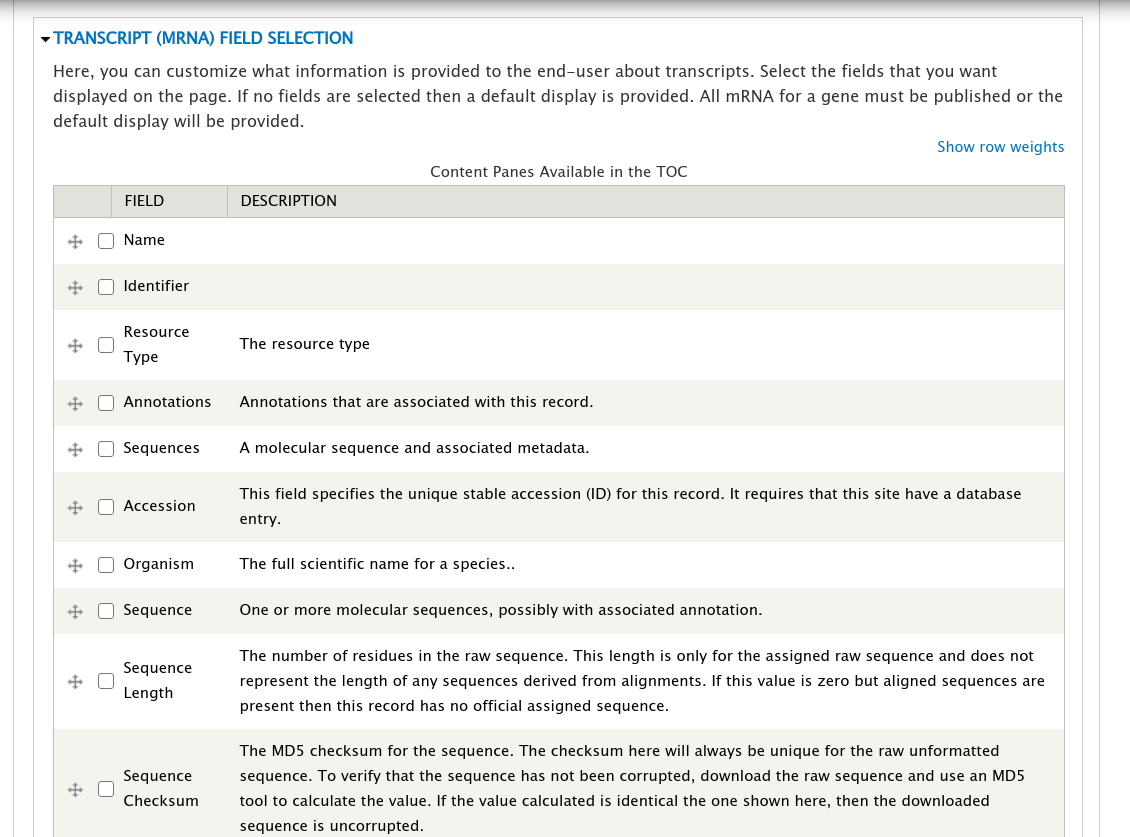
+Open the field set titled **Transcript (mRNA) Field Selection** to view a table that lists all of the available fields for a transcript.
|
|
|
+
|
|
|
+.. image:: genomes_genes.8.png
|
|
|
+
|
|
|
+On this page you can check the boxes next to the field that you want to show for a transcript on the gene page. For this example, we will select the fields **Name**, **Identifier**, **Resource Type**, **Anotations**, and **Sequences** (they may not be in this order on your own site). You can control the order in which fields will be shown by dragging them using the crosshairs icon next to each one. Scroll to the bottom of the page and click the **Save Settings** button.
|
|
|
+
|
|
|
+Next return to the gene page, reload it, and click on the **Transcripts** link. Now you are provided a select box with the transcript names. When a transcript is selected, the pane below will populate with the fields that you selected when editing in the Transcript field.
|
|
|
+
|
|
|
+.. image:: genomes_genes.9.png
|
|
|
+
|
|
|
+You can return to the Transcript field edit page under the Gene content type at any time to add, remove or change the order of fields that appear for the transcript.
|
|
|
+
|
|
|
+.. note::
|
|
|
+ Transcripts on a gene page can only be customized if all of them are published. If not, the default table listing is shown.
|