|
|
@@ -0,0 +1,181 @@
|
|
|
+/* override table width restrictions
|
|
|
+See: https://rackerlabs.github.io/docs-rackspace/tools/rtd-tables.html
|
|
|
+ */
|
|
|
+@media screen and (min-width: 767px) {
|
|
|
+
|
|
|
+ .wy-table-responsive table td {
|
|
|
+ white-space: normal !important;
|
|
|
+ }
|
|
|
+
|
|
|
+ .wy-table-responsive {
|
|
|
+ overflow: visible !important;
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
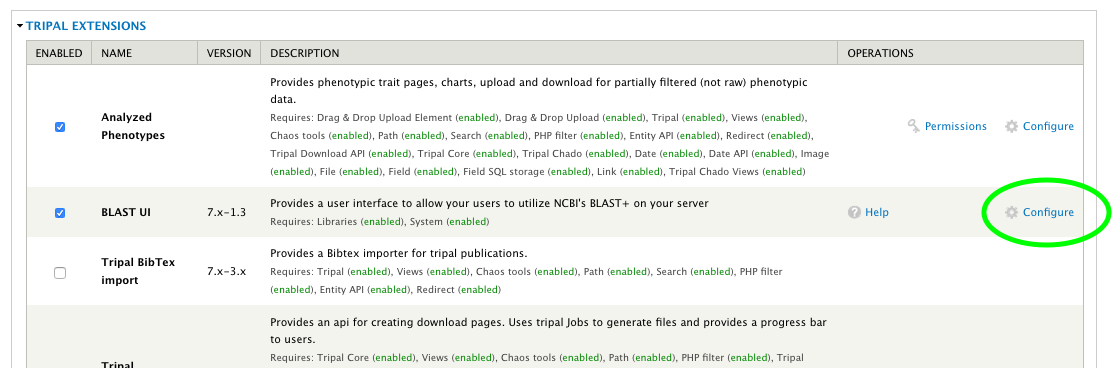
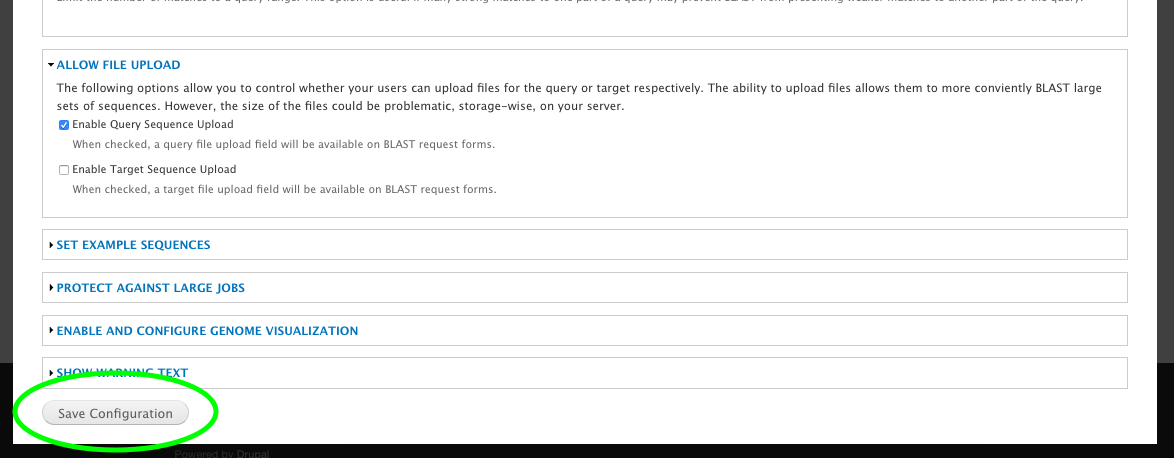
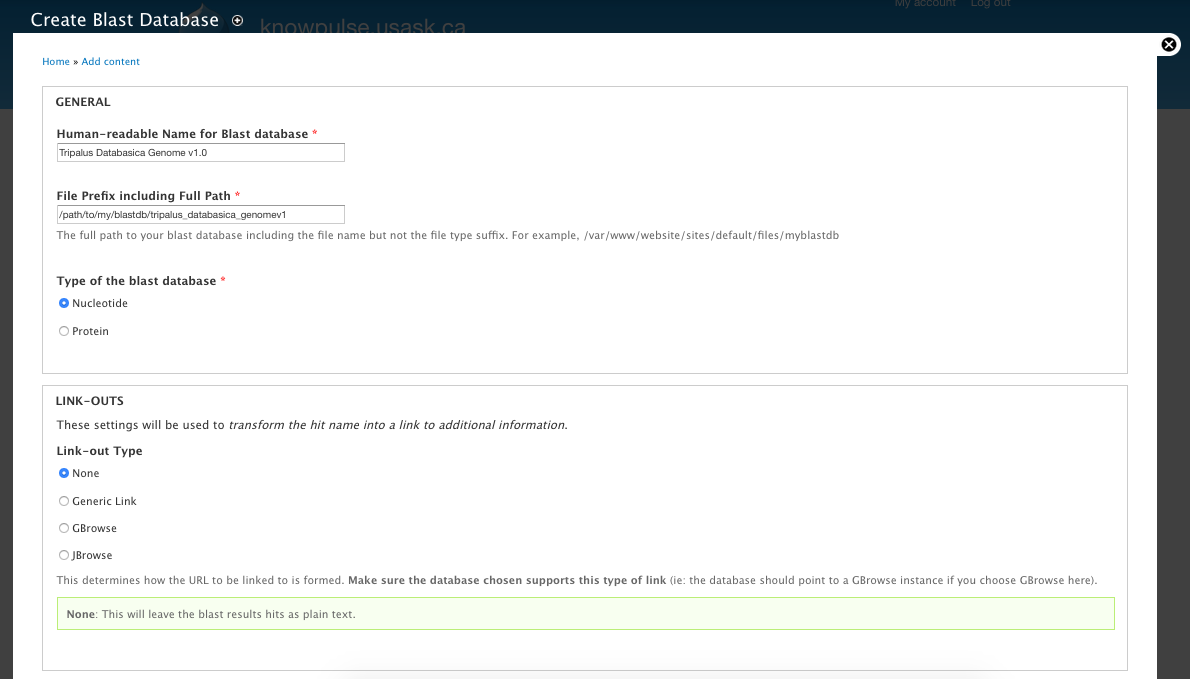
+ * Buttons on Extension Module pages.
|
|
|
+ */
|
|
|
+#administrative .section p:last-child a,
|
|
|
+ #analysis-annotation .section p:last-child a,
|
|
|
+ #data-loading-collection .section p:last-child a,
|
|
|
+ #developer-tools .section p:last-child a,
|
|
|
+ #third-party-integration .section p:last-child a,
|
|
|
+ #searching .section p:last-child a,
|
|
|
+ #visualization-display .section p:last-child a {
|
|
|
+ font-family: 'Helvetica Neue', Helvetica, Arial, sans-serif;
|
|
|
+ display: inline-block;
|
|
|
+ vertical-align: middle;
|
|
|
+ text-align: center;
|
|
|
+ border-radius: 4px;
|
|
|
+ margin-bottom: 20px;
|
|
|
+ margin-right: 8px;
|
|
|
+ -webkit-font-smoothing: antialiased;
|
|
|
+ letter-spacing: 0.03em;
|
|
|
+ font-weight: 500;
|
|
|
+ line-height: 14px;
|
|
|
+ font-size: 14px;
|
|
|
+ padding: 9px 10px;
|
|
|
+ color: #666666;
|
|
|
+ background: #FFFFFF;
|
|
|
+ background: -webkit-linear-gradient(top, #FFFFFF, #f1f1f1);
|
|
|
+ border: 1px solid #c8c8c8;
|
|
|
+ box-shadow: 0 1px 0px #e9e9e9;
|
|
|
+ }
|
|
|
+ #administrative .section p:last-child a:hover,
|
|
|
+ #analysis-annotation .section p:last-child a:hover,
|
|
|
+ #data-loading-collection .section p:last-child a:hover,
|
|
|
+ #developer-tools .section p:last-child a:hover,
|
|
|
+ #third-party-integration .section p:last-child a:hover,
|
|
|
+ #searching .section p:last-child a:hover,
|
|
|
+ #visualization-display .section p:last-child a:hover {
|
|
|
+ background: -webkit-linear-gradient(top, #FFFFFF, #e6e6e6);
|
|
|
+}
|
|
|
+
|
|
|
+/**
|
|
|
+ * BRANDING
|
|
|
+ */
|
|
|
+
|
|
|
+/* Sidebar Title */
|
|
|
+.wy-side-nav-search {
|
|
|
+ background: none !important;
|
|
|
+}
|
|
|
+.wy-side-nav-search input[type="text"] {
|
|
|
+ border-color: black;
|
|
|
+}
|
|
|
+.wy-nav-side {
|
|
|
+ color: #CCCCCC !important;
|
|
|
+ background: #2D2D34 !important;
|
|
|
+ background-image: url("hexagon_pattern.png") !important;
|
|
|
+ background-repeat: repeat !important;
|
|
|
+ background: -webkit-linear-gradient(left, $light-grey , $gray-base) !important;
|
|
|
+ background: linear-gradient(to right, $light-grey , $gray-base) !important;
|
|
|
+}
|
|
|
+/* Logo */
|
|
|
+.fa-home::before, .icon-home::before {
|
|
|
+ content: url('small_logoonly.png')
|
|
|
+}
|
|
|
+/* Sidebar TOC */
|
|
|
+.wy-menu-vertical li.current {
|
|
|
+ color: #CCCCCC;
|
|
|
+ border-color: black;
|
|
|
+ background: rgba(0,0,0,0.5);
|
|
|
+}
|
|
|
+.wy-menu-vertical li.toctree-l1.current > a {
|
|
|
+ color: black;
|
|
|
+ border-color: whitesmoke;
|
|
|
+ background-color: whitesmoke;
|
|
|
+}
|
|
|
+.wy-menu-vertical li.toctree-l2.current > a,
|
|
|
+ .wy-menu-vertical li.toctree-l2.current li.toctree-l3 > a,
|
|
|
+ .wy-menu-vertical li.toctree-l3.current li.toctree-l4 > a {
|
|
|
+ color: #CCCCCC;
|
|
|
+ border-color: black;
|
|
|
+ background-color: rgba(0,0,0,0.05);
|
|
|
+}
|
|
|
+.wy-menu-vertical li.current a:hover,
|
|
|
+ .wy-menu-vertical li.toctree-l1.current > a:hover,
|
|
|
+ .wy-menu-vertical li.toctree-l2.current > a:hover,
|
|
|
+ .wy-menu-vertical li.toctree-l2.current li.toctree-l3 > a:hover,
|
|
|
+ .wy-menu-vertical li.toctree-l3.current li.toctree-l4 > a:hover {
|
|
|
+ color: white;
|
|
|
+ border-color: black;
|
|
|
+ font-weight: bold;
|
|
|
+ background-color: rgba(0,0,0);
|
|
|
+}
|
|
|
+.wy-menu-vertical li.toctree-l1 a,
|
|
|
+ .wy-menu-vertical li.toctree-l2 a,
|
|
|
+ .wy-menu-vertical li.toctree-l3 a,
|
|
|
+ .wy-menu-vertical li.toctree-l4 a {
|
|
|
+ color: #CCCCCC;
|
|
|
+ border-color: black;
|
|
|
+}
|
|
|
+/* Main Content */
|
|
|
+.wy-nav-content-wrap {
|
|
|
+ background: whitesmoke;
|
|
|
+}
|
|
|
+a, a:hover {
|
|
|
+ color: #990000;
|
|
|
+}
|
|
|
+a:visited {
|
|
|
+ color: #cc0000;
|
|
|
+}
|
|
|
+a.icon-home:visited {
|
|
|
+ color: #ffffff;
|
|
|
+}
|
|
|
+/* Code blocks */
|
|
|
+.highlight {
|
|
|
+ background: #e1f0db;
|
|
|
+}
|
|
|
+/* Default Notes */
|
|
|
+.wy-alert.wy-alert-info,
|
|
|
+ .rst-content .note,
|
|
|
+ .rst-content .wy-alert-info.attention,
|
|
|
+ .rst-content .wy-alert-info.caution,
|
|
|
+ .rst-content .wy-alert-info.danger,
|
|
|
+ .rst-content .wy-alert-info.error,
|
|
|
+ .rst-content .wy-alert-info.hint,
|
|
|
+ .rst-content .wy-alert-info.important,
|
|
|
+ .rst-content .wy-alert-info.tip,
|
|
|
+ .rst-content .wy-alert-info.warning,
|
|
|
+ .rst-content .seealso, .rst-content
|
|
|
+ .wy-alert-info.admonition-todo,
|
|
|
+ .rst-content .wy-alert-info.admonition {
|
|
|
+ background: #d9d9d9;
|
|
|
+}
|
|
|
+.wy-alert.wy-alert-info
|
|
|
+ .wy-alert-title,
|
|
|
+ .rst-content .note .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.attention .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.caution .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.danger .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.error .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.hint .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.important .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.tip .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.warning .wy-alert-title,
|
|
|
+ .rst-content .seealso .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.admonition-todo .wy-alert-title,
|
|
|
+ .rst-content .wy-alert-info.admonition .wy-alert-title,
|
|
|
+ .wy-alert.wy-alert-info .rst-content .admonition-title,
|
|
|
+ .rst-content .wy-alert.wy-alert-info .admonition-title,
|
|
|
+ .rst-content .note .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.attention .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.caution .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.danger .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.error .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.hint .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.important .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.tip .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.warning .admonition-title,
|
|
|
+ .rst-content .seealso .admonition-title, .rst-content
|
|
|
+ .wy-alert-info.admonition-todo .admonition-title,
|
|
|
+ .rst-content .wy-alert-info.admonition .admonition-title {
|
|
|
+ background: #737373;
|
|
|
+}
|
|
|
+/* Note Warnings */
|
|
|
+.wy-alert.wy-alert-warning .wy-alert-title, .rst-content .wy-alert-warning.note .wy-alert-title, .rst-content .attention .wy-alert-title, .rst-content .caution .wy-alert-title, .rst-content .wy-alert-warning.danger .wy-alert-title, .rst-content .wy-alert-warning.error .wy-alert-title, .rst-content .wy-alert-warning.hint .wy-alert-title, .rst-content .wy-alert-warning.important .wy-alert-title, .rst-content .wy-alert-warning.tip .wy-alert-title, .rst-content .warning .wy-alert-title, .rst-content .wy-alert-warning.seealso .wy-alert-title, .rst-content .admonition-todo .wy-alert-title, .rst-content .wy-alert-warning.admonition .wy-alert-title, .wy-alert.wy-alert-warning .rst-content .admonition-title, .rst-content .wy-alert.wy-alert-warning .admonition-title, .rst-content .wy-alert-warning.note .admonition-title, .rst-content .attention .admonition-title, .rst-content .caution .admonition-title, .rst-content .wy-alert-warning.danger .admonition-title, .rst-content .wy-alert-warning.error .admonition-title, .rst-content .wy-alert-warning.hint .admonition-title, .rst-content .wy-alert-warning.important .admonition-title, .rst-content .wy-alert-warning.tip .admonition-title, .rst-content .warning .admonition-title, .rst-content .wy-alert-warning.seealso .admonition-title, .rst-content .admonition-todo .admonition-title, .rst-content .wy-alert-warning.admonition .admonition-title {
|
|
|
+ background: #e6b800;
|
|
|
+}
|
|
|
+.wy-alert.wy-alert-warning, .rst-content .wy-alert-warning.note, .rst-content .attention, .rst-content .caution, .rst-content .wy-alert-warning.danger, .rst-content .wy-alert-warning.error, .rst-content .wy-alert-warning.hint, .rst-content .wy-alert-warning.important, .rst-content .wy-alert-warning.tip, .rst-content .warning, .rst-content .wy-alert-warning.seealso, .rst-content .admonition-todo, .rst-content .wy-alert-warning.admonition {
|
|
|
+ background: #FcFde6;
|
|
|
+}
|